Figure 1.
Biallelic Integration of FP-SNP Pairs Enable Deterministic Genotype Outcomes
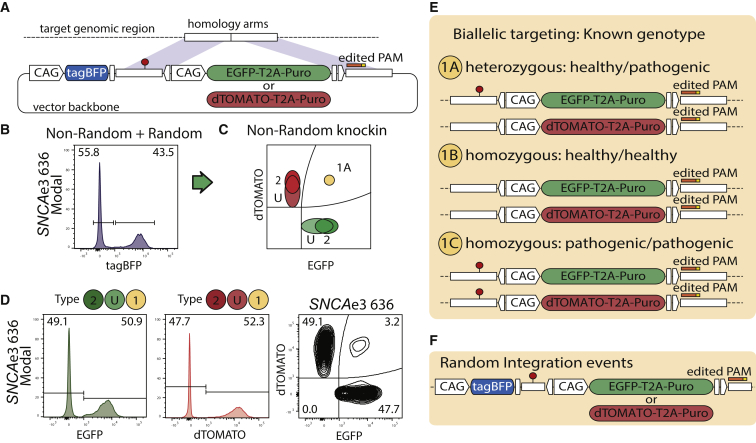
(A) Donor vectors contain a PSM expressing EGFP or dTOMATO, and an NSM expressing tagBFP. PSMs contain puromycin resistance gene (Puro).
(B) Representative example of SNCAe3 polyclone 636. Random integration tagBFPpos cells are excluded from the correctly edited on-target cells tagBFPneg.
(C) Theoretical distribution of populations for non-random outcomes.
(D) Representative example of SNCAe3. On-target cells include homozygous populations, EGFPpos/EGFPpos or dTOMATOpos/dTOMATOpos (type 2), and heterozygous populations of undefined second-allele state EGFPpos/WT-NHEJ or dTOMATOpos/WT-NHEJ (type U). WT, wild-type.
(E) Outcomes of the derived population are defined according to the donor vector design.
(F) The tagBFP NSM allows removal of random integration events, assisting in the derivation of defined outcomes.