Figure 1.
AHR Tagging Strategy and Functional Validation of the Tagged Protein
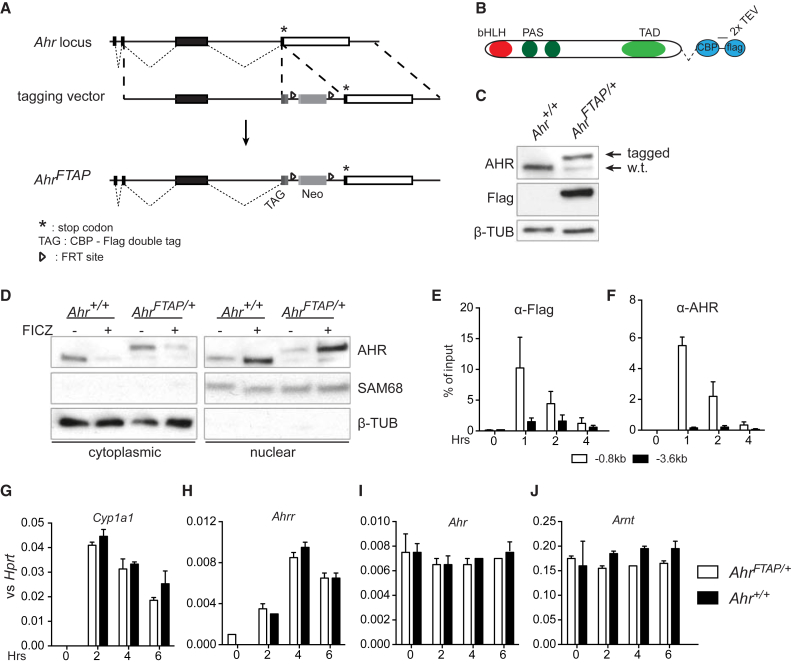
(A) Graphic representation of the 3′ end of the Ahr locus depicting the knockin strategy for c-terminal tagging of the AHR protein, showing the wild-type Ahr locus, the targeting vector, and the resulting AhrFTAP allele. STOP codon is marked by an asterisk, coding sequences represented as black boxes, and 3′ UTRs as open boxes. Small dashed lines join splice junctions, and larger dashed lines mark homologous regions.
(B) The protein product of the AhrFTAP allele showing the full-length AHR protein and its domains fused to the tag shown in blue. bHLH, basic-helix-loop-helix; PAS, period-ARNT-sim domain; TAD, transcription activation domain.
(C) Western blot of whole-cell lysate from the paternal Ahr+/+ and the targeted AhrFTAP/+ ESCs.
(D) Western blot of cytoplasmic and nuclear fractions from Ahr+/+ and AhrFTAP/+ ESCs treated with vehicle or FICZ for 1 hr using antibodies against the indicated proteins. SAM68 and tubulin beta mark nuclear or cytoplasmic localization, respectively, and also serve as loading controls. Western blots in (C and D) are representative of at least two experiments.
(E and F) Chromatin immunoprecipitation using antibodies against Flag or AHR on chromatin extracted from AhrFTAP/+ ESCs treated with vehicle or FICZ for the indicated time points. Immunoprecipitated DNA was detected with primers against the Cyp1a1 dioxin response element at −0.8 kb from the transcription start site of the gene (white bars) or an irrelevant region further upstream at −3.6 kb (black bars) as negative control. Results are represented as percentage of input DNA and shown as averages +SEM from three experiments.
(G–J) RT-qPCR on RNA from Ahr+/+ (black bars) and AhrFTAP/+ (white bars) ESCs for the indicated genes. Data expressed relative to Hprt abundance and shown as averages +SEM from two experiments.