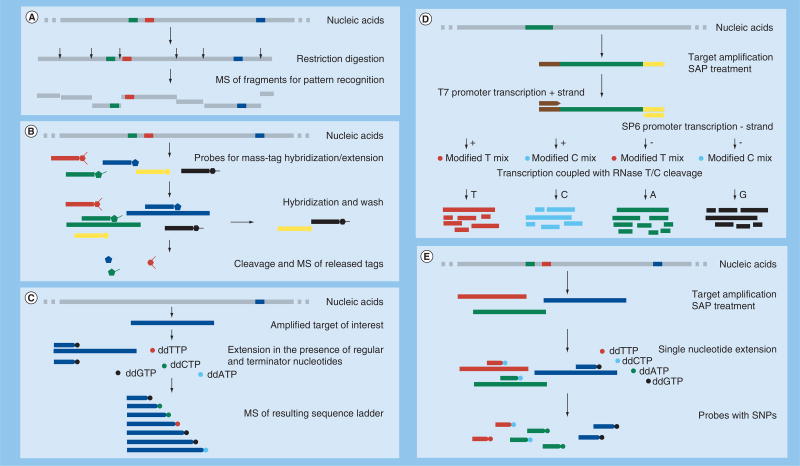
Figure 3. Biochemical processes for comprehensive molecular analysis of specific gene targets as applied to analysis by mass spectrometry.
(A) Restriction fragment length polymorphism – the complexity of large target molecules is reduced by enzymatic cleavage at predetermined sites, the resulting small molecules are analyzed by MS and the obtained mass patterns are used to infer organism identification. (B) Mass-tag multiplexing – each target is detected by a probe tagged by a cleavable small molecule, unused probes are removed/washed, the tags of the used probes are cleaved and used for detection by MS; detected tags indicate presence of the target of interest. (C) Generation of sequence ladder in the presence of one specific primer, regular nucleotides, dideoxynucleotides and polymerase, resulting in random termination of the fragments at any and all positions, allowing the discrimination of two fragments by one added nucleotide. (D) Sequencing by transcription with T6 and SP6 polymerases, coupled to RNase A single base-specific cleavage. The process queries both strands of each amplicon of interest. The resulting mass fingerprint contains information about all four nucleotides and is used to derive the fragment’s sequence, genotype and heterogeneity and to discover new mutations by in silico pattern comparison to a comprehensive reference set. (E) SNP identification by multiplex PCR. Each product is then queried by a specific probe designed immediately upstream from a SNP of interest and then extended in the presence of ddNTP mix. The resulting extended probes present a mass signature that identifies the SNP sequence. Depending on the assay design (e.g., T5000) the multiplex product could be used directly for MS with the resulting mass patterns used for organism identification.
MS: Mass spectrometry; SAP: Shrimp alkaline phosphatase.