Figure 3. Determinants of nucleosome positioning.
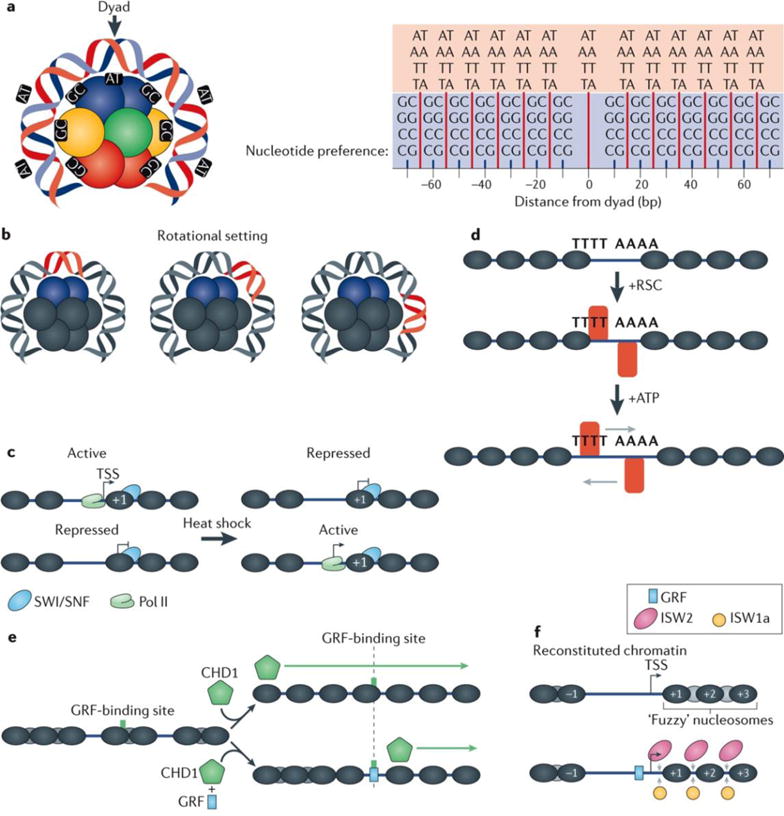
a | Nucleosomal DNA, particularly at +1 and −1 positions with respect to transcription start sites (TSSs), is modestly enriched with AA, TT, AT, and TA dinucleotides at a 10 bp periodicity and GG, CC, GC, and CG dinucleotides offset by 5 bp from those dinucleotides and also possessing a 10 bp periodicity36,38,50,54,74. These periodicities result in the formation of preferred bends in DNA, thereby defining how DNA wraps around the histone octamer. Point 0 represents the dyad with preference for AA, TT, AT and TA dinucleotides, and preference in 10 bp periodicity at 15, 25, 35, 45, 55 and 65 bp from the dyad are represented by the red lines. The GG, CC, GC and CG dinucleotide preferences are represented by the short blue lines at points 10, 20, 30, 40, 50, 60 and 70 bp from the dyad. b | The position of DNA relative to the histone octamer defines its rotational setting. DNA sequences possessing the nucleotide periodicities shown in part a possess a strong rotational setting, which means that they typically wrap around the octamer in a preferred orientation. However, for a single rotational setting, multiple translational settings can exist, typically in 10 bp intervals, reflecting the nucleotide preference periodicities in the DNA. c | The switch/sucrose non-fermenting (SWI/SNF) chromatin remodelling complex helps to regulate various cellular processes, including stress responses, such as heat shock. It may do so by altering the accessibility of promoter DNA to regulatory proteins and the transcription machinery (contributing to both activation and repression of transcription)14,17. d | The highly abundant and essential remodelling the structure of chromatin (RSC) complex acts on poly(dA:dT) tracts, which are common in Saccharomyces cerevisiae promoters. RSC regulates the size of the nucleosome-free region in these promoters by repositioning and/or evicting nucleosomes located near poly(dA:dT) tracts in an ATP-dependent manner18,90,91. e | The chromodomain helicase DNA binding 1 (CHD1) remodeller produces regularly spaced nucleosome arrays in vitro using ATP95. The targeting of nucleosome remodelling activity of CHD1 is suspected to be linked to transcription98,99, which explains its apparent lack of DNA-binding specificity in genome-wide in vitro nucleosome assembly assays18. f | In S. cerevisiae the imitation SWI (ISWI) family of chromatin remodellers are enriched at the 5′ ends of gene bodies25. In vitro experiments have revealed that ISW1a and ISW2 are crucial for establishing proper in vivo nucleosome spacing18. Spacing is further improved with the addition of general transcription factors (GRFs), which supports the predicted role of GRFs as anchor points, or boundary elements, to guide the directionality for ISW1a112. See also part e. Pol II, RNA polymerase II.
