Abstract
Peptidoglycan recognition proteins (PGRPs) and commensal microbes mediate pathogen infection outcomes in insect disease vectors. Although PGRP-LD is retained in multiple vectors, its role in host defense remains elusive. Here we report that Anopheles stephensi PGRP-LD protects the vector from malaria parasite infection by regulating gut homeostasis. Specifically, knock down of PGRP-LD (dsLD) increased susceptibility to Plasmodium berghei infection, decreased the abundance of gut microbiota and changed their spatial distribution. This outcome resulted from a change in the structural integrity of the peritrophic matrix (PM), which is a chitinous and proteinaceous barrier that lines the midgut lumen. Reduction of microbiota in dsLD mosquitoes due to the upregulation of immune effectors led to dysregulation of PM genes and PM fragmentation. Elimination of gut microbiota in antibiotic treated mosquitoes (Abx) led to PM loss and increased vectorial competence. Recolonization of Abx mosquitoes with indigenous Enterobacter sp. restored PM integrity and decreased mosquito vectorial capacity. Silencing PGRP-LD in mosquitoes without PM didn’t influence their vector competence. Our results indicate that PGPR-LD protects the gut microbiota by preventing hyper-immunity, which in turn promotes PM structurally integrity. The intact PM plays a key role in limiting P. berghei infection.
Author summary
Malaria parasites must overcome several obstacles to complete their development in mosquito. Understanding the interactions between parasites and mosquitoes will provide potential targets to control malaria transmission. PGRP-LD is a peptidoglycan recognition protein, of which limit information is available in insects. Here we show that A. stephensi PGRP-LD mediates malaria parasite infection outcomes by influencing homeostasis of the gut microbiota. Reduction of the gut microbiota density, resulting from upregulation of immune activities in PGRP-LD knock down mosquitoes, changes expression of PM genes and causes PM fragmentation. The compromised PM leads to increasing susceptibility to parasite infection. We also discovered that the PM is lost in mosquitoes in which the gut microbiota is removed by antibiotic treatment. Knock down of PGRP-LD in these mosquitoes doesn’t increase their vector competence. Altogether, these results indicate that capacity of Anopheles mosquito to transmit parasites is determined by a finely tuned balance between host immunity, gut microbiota and peritrophic matrix. PGRP-LD is a key mediator in regulating this balance. Our results expand knowledge on interactions between immune system, gut microbiota and Plasmodium, and will shed light on equivalent processes in other disease transmitting vectors.
Introduction
Malaria is caused by parasites from the genus Plasmodium. The disease kills over 500,000 people annually, most of which are children under the age of 5 [1]. In order to transmit between humans, Plasmodium must overcome several obstacles to complete its development in Anopheles mosquitoes [2–4]. The peritrophic matrix (PM), and immuno-competent midgut epithelial cells, are two barriers that interfere with parasite transmission through their mosquito vector. The PM is non-cellular and composed of chitin fibrils and chitin-binding proteins. The structure lines the midgut lumen and wraps the food bolus within the endoperitrophic space, thus protecting the epithelium from abrasive food particles and enteric pathogens [5]. The tight junctions between midgut epithelial cells form another contiguous barrier against parasite invasion [6]. Midgut epithelial cells invaded by Plasmodium undergo apoptosis and are replaced by new cells. This rapid turnover not only maintains the integrity of the epithelium, but also clears invading parasites [7]. In addition to overcoming physical barriers present in the mosquito midgut, epithelial cells in this environment also present robust cellular and humoral immunity [3]. This activity includes the synthesis of antimicrobial peptides (AMPs), reactive oxygen species (ROS) and nitric oxide (NO), all of which contribute to parasite clearance [4]. Anopheles mosquitoes have 3 types of hemocytes: granulocytes, oenocytoids and prohemocytes [3]. These cells eliminate pathogens via phagocytosis and encapsulation. Hemocytes are also important in Plasmodium-mediated immune memory, which enhances the mosquito’s ability to clear parasites upon reinfection [8]. In addition, complement like protein TEP1 by forming TEP1/LRIM1/APL1C complex, is another key systemic antiplasmodial immune mechanism that recognizes and eliminates Plasmodium ookinetes in the midgut [3]. Three major immune signaling pathways, Toll, IMD (Immune Deficiency) and JAK/STAT, are critical mediators of malaria infection dynamics in Anopheles mosquitoes [3].
Peptidoglycan recognition proteins (PGRP) are pattern recognition molecules that function as receptors and regulators of the Toll and IMD signaling pathways [9]. Anopheles has 7 PGRP genes, 4 in the Long subfamily (including PGRP-LA, -LB, -LC and -LD) and 3 in the short subfamily (PGRP-S1, -S2 and –S3) [10]. Anopheles PGRP-LC is a receptor of the Immune Deficiency (Imd) pathway that is responsible for triggering synthesis of downstream effector molecules [11]. Knock down of PGRP-LC results in increasing susceptibility to Plasmodium infection. PGRP-LA, another receptor of the Imd pathway, protects A. coluzzii from Plasmodium infection in a manner similar to that of PGRP-LC [12]. PGRP-LB, a negative regulator of the Imd pathway, has a dual role in Anopheles mosquitoes, facilitating parasite infection and protecting natural gut bacteria [12,13]. However, mechanisms of other PGRPs in response to parasite infection are still inadequate.
The gut microbiota is another important factor that strongly influences vector competence [14]. Interactions between enteric bacteria and the mosquito immune system help to maintain gut homeostasis and protect mosquitoes from pathogens infection [13,15–17]. In the absence of gut microbes, Anopheles become highly susceptible to Plasmodium infection. Co-feeding parasites with bacteria restores resistance to parasite infection in mosquitoes previously treated with antibiotics to remove their indigenous microbiota. Gut microbes also induce expression of several immune molecules, including antimicrobial peptides and pattern recognition receptors [13], and enhance vector refractoriness by promoting hemocyte differentiation [8]. Some residential bacteria, including Enterobacter and Chromobacterium isolated from field mosquitoes, directly inhibit parasite infection by secreting secondary metabolites such as reactive oxygen species [15,18].
In this study, we examined the function of PGRP-LD in A. stephensi and found that this receptor protects the mosquito against Plasmodium infection. PGRP-LD helped maintain homeostasis of the mosquito gut microbiota by negatively regulating innate immune responses. The healthy microbiota in turn contributed to the integrity of PM, and the intact PM enhanced Anopheles resistance to malaria parasites. Our results suggest that a finely tuned balance between the immune system, gut microbes and the PM is key to determining the capacity of mosquitoes to transmit malaria.
Results
PGRP-LD helps to defend against parasite infection
The putative Anopheles stephensi PGRP-LD is 42 kD transmembrane protein with 77% identity to Anopheles gambiae PGRP-LD. Sequence analysis indicates that it has a peptidoglycan-binding domain. However, the putative protein lacks most of the residues essential for PGN binding and catalytic activity, which are well characterized domains of Drosophila PGRPs (S1 Fig).
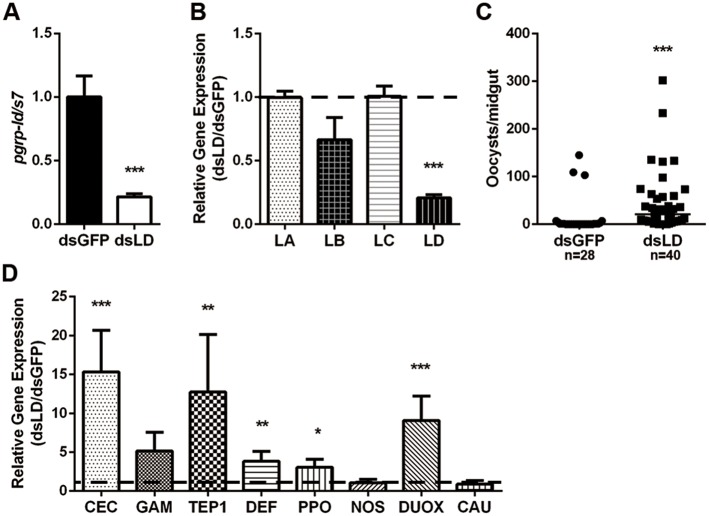
To investigate the role of PGRP-LD in parasites defense, we knocked down its expression in vivo via microinjection of gene-specific double stranded RNA and then analyzed the susceptibility of treated mosquitoes to infection with P. berghei. The level of pgrp-ld was reduced by approximately 67% 2-days post dsRNA treatment compared to dsGFP controls (Fig 1A), and we observed no significant cross reactivity with other long PGRPs, including PGRP-LA, -LB, -LC (Fig 1B). Knock down of pgrp-ld didn’t influence the survival rate of mosquitoes (S2 Fig). However, reduced PGRP-LD (dsLD) resulted in a significant increase in the number of oocysts from 0 in dsGFP to 31 in dsLD mosquitoes (Fig 1C).
Fig 1. Influence of PGRP-D on P. berghei infection in A. stephensi.
(A) pgrp-ld silencing efficiency and specificity (B) in mosquitoes. Expression level of pgrp-ld was normalized to A. stephensi s7. Relative expression level of pgrp-ld in dsLD mosquitoes was normalized to the gene’s expression in dsGFP controls. (C) Oocyst number in dsRNA treated mosquitoes. Median oocysts number is indicated by the horizontal black bar. Each dot represents an individual mosquito. (D) Relative gene expression levels in dsRNA treated mosquitoes 26hr post parasite infection. (A, B and D) Error bars indicate standard error (n = 10). Significance was determined by Student’s-T test in (A), (B) and (D), by Mann-Whitney test in (C). *, P<0.05, **, P<0.01, ***, P<0.001. Results from one of three independent experiments are shown.
As PGRPs play important roles in activation and regulation of immune responses, we hypothesized that increased susceptibility of dsLD mosquitoes to parasites infection might resulted from the dysregulation of innate immune responses [9]. To address this question we next analyzed expression of 8 immune genes in dsLD and dsGFP treated mosquitoes 26hr post parasite challenge. The genes we investigated encoded 3 antimicrobial peptides (Cecropin, Gambicin and Defensin), 1 negative regulator of IMD signaling pathway (Caudal) and 4 proteins related to cellular and epithelial immune responses (TEP1, PPO, NOS and DUOX) [3,19]. Interestingly, most of the effector encoding genes, including cecropin, defensin, tep1, ppo and duox, were significantly upregulated in response to parasite challenge (Fig 1D). However, these induced effectors did not control parasite infection outcomes. This finding suggests a discrepancy exists between increased susceptibility to parasites and enhanced expression of immune genes in the absence of PGRP-LD.
PGRP-LD contributes to the homeostasis of gut microbiota
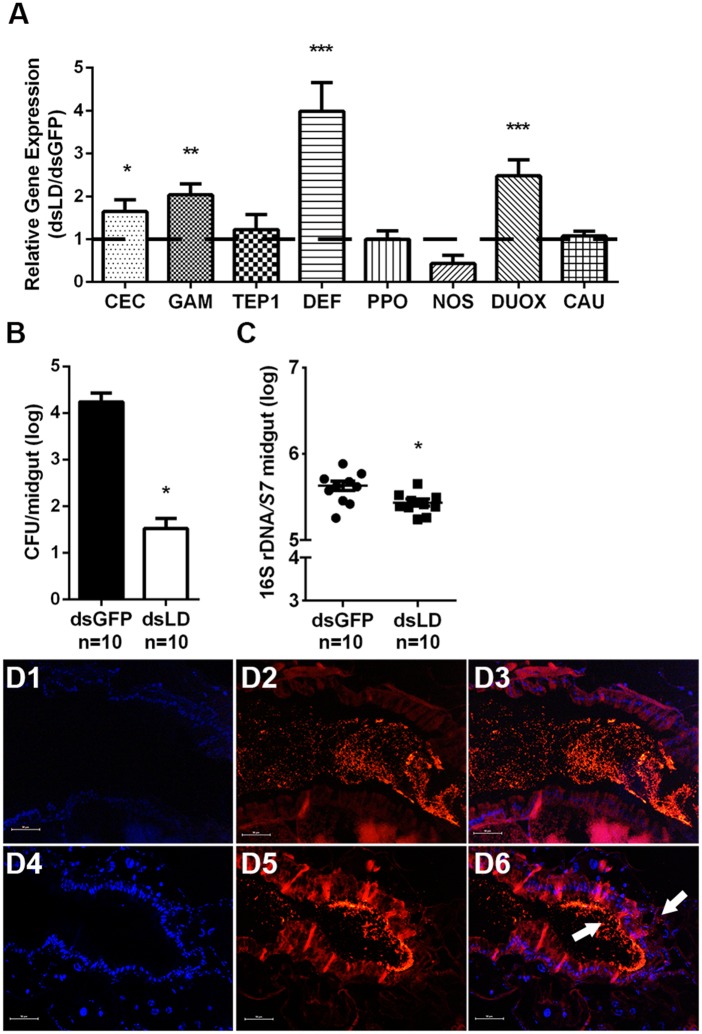
We next examined if pgrp-ld similarly regulated immune responses in mosquitoes prior to blood meal. The same 8 genes were expressed in mosquitoes fed only on sugar. As expected, 4 of these genes (cecropin, gambicin and defensin, and duox) were upregulated in dsLD treated mosquitoes, while tep1, ppo, nos and caudal expression remained unchanged (Fig 2A). As both antimicrobial peptides and ROS present bactericidal activities, we next examined if over-activated immune responses exerted an influence on microbiota homeostasis [20,21]. Bacterial load of both culturable and unculturable bacteria were measured in dsLD mosquitoes before consumption of a blood meal. In agreement with our hypothesis, knock down of pgrp-ld resulted in an ~500 times reduction of culturable microbes such that dsGFP individuals housed average 1.7X104 CFU/midgut, while dsLD individuals housed average 3.3X101 CFU/midgut (Fig 2B). Similarly, the 16s rRNA gene copy number was significantly lower in dsLD compared to dsGFP mosquitoes (Fig 2C). We next analyzed if community structure of the gut microbiota was influenced in the absence of PGRP-LD. Midguts of dsRNA treated mosquitoes were dissected and bacterial community structure was determined by 16S rRNA next generation sequencing. No significant difference in taxonomic structure was observed between microbial communities in dsGFP and dsLD mosquitoes (S3 Fig). These results indicate that over-activated immune responses in the presence of reduced pgrp-ld expression leads to a reduction in the number of residential bacteria, without influencing the taxonomic composition of the gut microbial community. Thus, PGRP-LD helps to protect commensal bacteria by preventing the overactivation of host immune responses.
Fig 2. Influence of PGRP-LD on the gut microbiota.
(A) Relative expression levels of immunity-related genes in dsRNA treated, sugar fed mosquitoes. Error bars indicate standard error (n = 10). Culturable (B) and total gut microbiota (C) density was measured. Error bars indicate standard error (n = 10). Significance was determined by Student’s-T test. *, P<0.05, **, P<0.01, ***, P<0.001. Results from one of two independent experiments are shown. (D) Microbiota localization in midguts of dsGFP (D1-D3) and dsLD (D4-D6) were analyzed by FISH using a universal bacterial 16S rRNA gene probe (red). Nuclei were stained with DAPI (blue). D1 and D4 show DAPI staining. D2 and D5 show staining with 16S rRNA probe. D3 and D6 show merged images. Arrows denote gut microbiota. Images are representative of three independent experiments. Scale bars, 50 μm.
In addition to investigating bacterial abundance and taxonomic composition, we also examined the spatial distribution of residential bacteria in dsLD mosquito midguts. Localization of residential bacteria in A. stephensi midguts was examined 48hr post blood meal, which is when cumulative population reaches its maximum density as determined by fluorescent in situ hybridization (FISH) using a universal 16s ribosomal RNA (rRNA) gene probe [22]. We observed a clear physical separation of gut microbiota and epithelium in dsGFP controls (Fig 2D1–2D3). However, dsLD treated mosquitoes exhibited a defect in spatial segregation, with increasing bacteria coming into direct contact with the gut epithelium, and even penetrating epithelial cells (Fig 2D4–2D6). Taken together, these results suggest that PGRP-LD helps to maintain the spatial homeostasis of gut microbes.
Peritrophic matrix is compromised in the absence of PGRP-LD
The PM, which is composed of chitin fibrils and glycoproteins, is a sheath like structure that lines the digestive tract of most insect midguts and prevents luminal contents from coming into direct contact with midgut epithelial cells [23,24]. Mosquitoes have type I PMs, the formation of which is triggered by ingestion of a blood meal [5]. We hypothesized that the microbial diffusion we observed in dsLD midguts may occur because these mosquitoes present a structurally compromised PM. We thus analyzed PM structure in dsLD and dsGFP mosquitoes by hematoxylin and eosin (H&E) and Periodic Acid Schiff (PAS) staining. A fully formed PM was visualized in dsGFP controls 48 hr post blood meal (Figs 3A1 and 3A2 and S4A1). Conversely, the PM of dsLD mosquitoes appeared fragmented (Figs 3A3 and 3A4 and S4A2). To further confirm the impaired PM structure in dsLD mosquitoes, dsRNA treated individuals were fed a blood meal supplemented with FITC-labelled dextran molecules (500 kDa). We observed dextran beads were restrained within the endoperitrophic space in dsGFP mosquitoes 48 hr post feeding (Fig 3B1). In contrast, we observed beads penetrating gut epithelial cells in dsLD mosquitoes, indicating that PM structure was compromised when pgrp-ld expression was experimentally reduced (Fig 3B2). We next examined if impaired PM structure was due to the dys-regulation of PM genes. We monitored expression of 2 peritrophin genes (peritrophin1 and 14), and 2 chitinases (chitinaseA and chitinaseB), all of which are involved in the PM formation and degeneration, in dsRNA treated mosquitoes 24hr and 48hr post blood feeding [25,26]. When pgrp-ld expression was knocked down, the 2 chitinases were upregulated 24 hr post blood meal, followed by a significant downregulation 48hr post blood meal comparing to dsGFP controls. Expression of peritophin 1 was lower at both time points, with a significant reduction 48hr post blood meal (Fig 3C). These data reinforce our hypothesis that the compromised PM in dsLD mosquitoes is due to the dysregulation of PM associated genes. Taken together, these results suggest that PGRP-LD plays a role in maintaining PM structural integrity in the gut of A. stephensi.
Fig 3. PM structure in dsRNA treated mosquitoes.
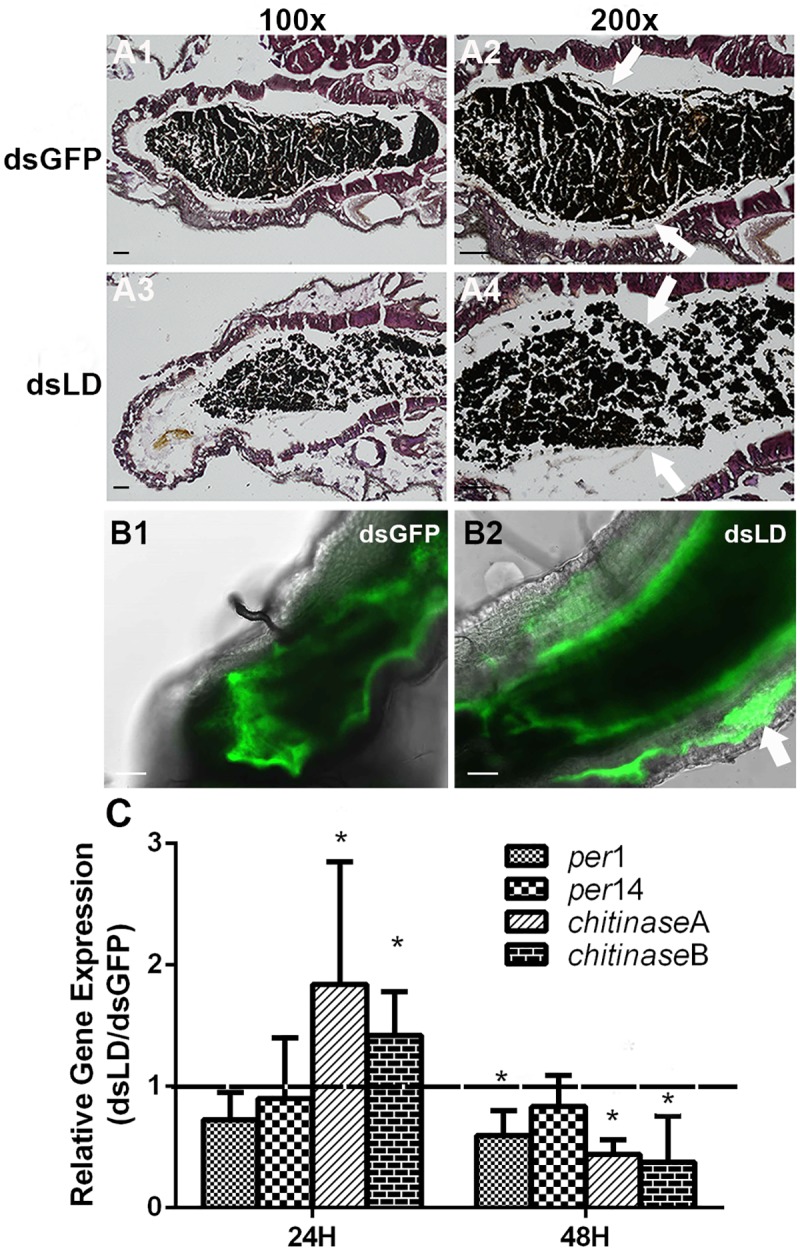
(A) PM structure was observed by H&E staining in dsGFP (A1 and A2) and dsLD (A3 and A4) mosquitoes at 100X (A1 and A3) and 200X (A2 and A4) magnification. Arrows denote the PM. Scale bars, 50 μm. (B) Dextran-feeding assay in dsRNA treated mosquitoes. The FITC signal is retained in the lumen of dsGFP control mosquitoes, which indicates that the dextran beads are contained within the PM (B1). The FITC signal is observed within gut epithelial cells (indicated by arrow) in dsLD mosquitoes, indicating that the beads can cross the PM (B2). Scale bars, 50 μm. Images are representative of at least two independent experiments. (C) PM gene expression in dsRNA treated mosquitoes. Relative gene expression level in dsRNA treated mosquitoes 24hr and 48 hr post blood meal. Error bars indicate standard error (n = 10).
Gut microbiota promotes structural integrity of peritrophic matrix
Gut microbes promote PM structural integrity [27–29]. Because gut microbe abundance was significantly reduced in dsLD mosquitoes, impaired PM structure in these mosquitoes may be due to gut dysbiosis. We next analyzed if resident microbes impact PM structure in A. stephensi. We again examined the structure of the PM in both normal and antibiotic treated mosquitoes (Abx) 48-hour post blood meal by H&E staining. Antibiotic treatment cleared the majority of native gut bacteria (S5 Fig). Furthermore, an intact PM was observed in guts of normal mosquitoes, which contained the blood bolus within the endoperitrophic space (Fig 4A and 4B). In contrast, when the gut microbiota was removed, no PM was observed and blood was dispersed within the entire gut lumen (Fig 4C and 4D). We again analyzed expression of the same 4 PM genes and found similar expression profiles as in dsRNA treated mosquitoes, with a decrease in the expression of peritrophin1 and 14 and an initial increase of PM digesting chitinases 24 hr post blood meal in antibiotic treated mosquitoes (Fig 4E). Thus, gut microbes may play a role in regulating expression of PM genes, thereby maintaining PM structural integrity.
Fig 4. PM structural integrity in normal and antibiotic treated mosquitoes.
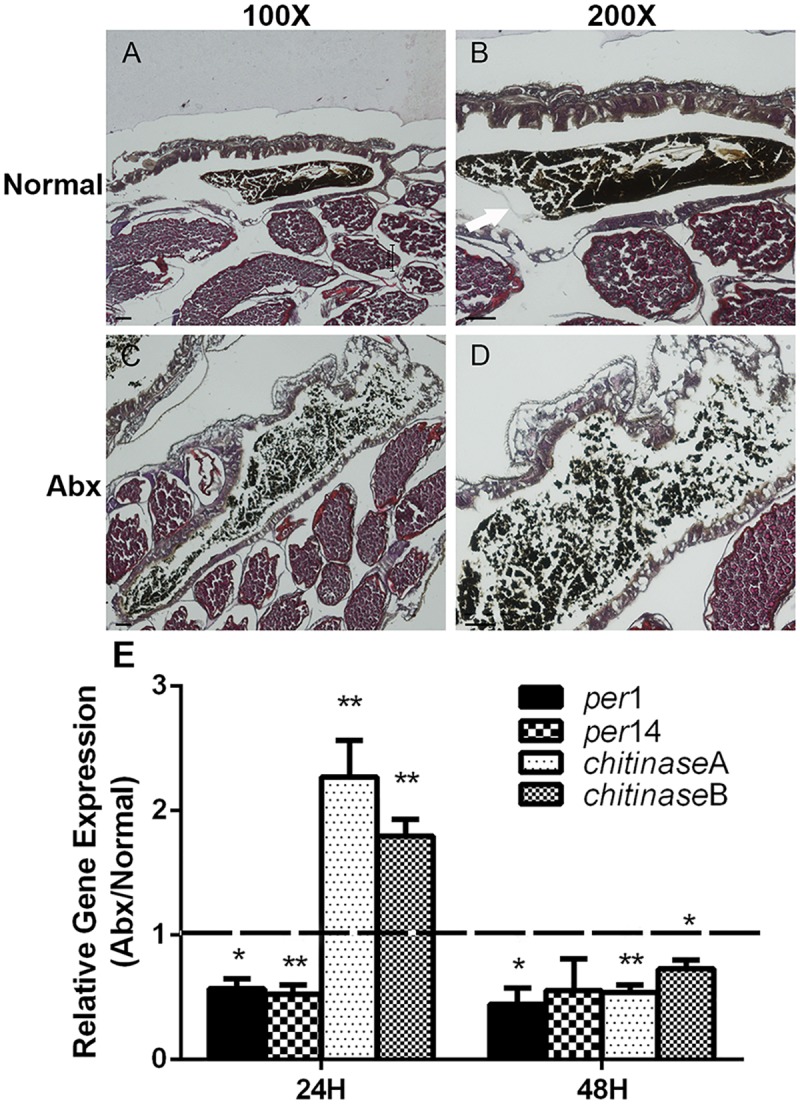
PM structure was observed by H&E staining in normal (A and B) and antibiotic treated mosquitoes (C and D) at 100X (A and C) and 200X (B and D) magnification. Arrows denote the PM. Images are representative of three independent experiments. Scale bars, 50 μm. (E) Quantification of PM related gene expression in normal and antibiotic treated mosquitoes 24hr and 48 hr post blood meal. Error bars indicate standard error (n = 10).
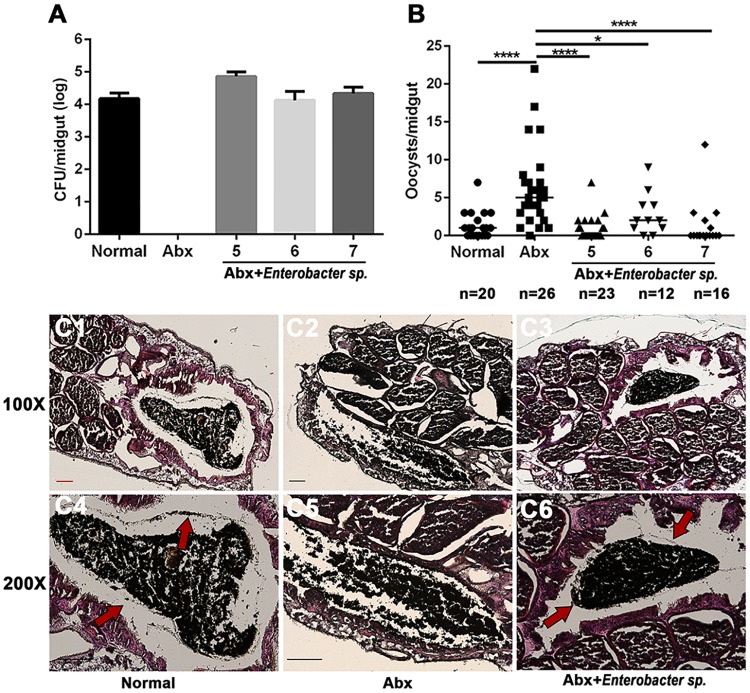
To further analyze the functional association between gut microbes and PM structure, we colonized guts of antibiotic treated mosquitoes with Enterobacter sp. (three different doses, 1X105/ml, 106/ml and 107/ml 1.5% sugar solution) prior to administering a blood meal. As Enterobacter cloacae is able to inhibit Plasmodium infection in A. stephensi [30], we then examined if Enterobacter sp. isolated from our mosquito colony were able to inhibit parasite colonization. Two days post-inoculation, each concentration reached an average density of 7.2X104/ midgut, 1.3X104/midgut and 2.2X104/midgut, respectively, which is comparable to that found indigenously in normal mosquitoes (1.5X104 CFU/midgut) (Fig 5A). We next examined the infection rate in these mosquitoes and found that increasing susceptibility to P. berghei infection was rescued to normal levels when Abx treated mosquitoes were re-colonized with all three Enterobacter concentrations (Fig 5B). Because no difference in infection rate was observed in the three inoculation concentrations, the PM of mosquitoes recolonized with 1X105/ml Enterobacter sp. was stained with H&E and PAS 2-day post blood meal. Clear PM structures were observed in both normal mosquitoes (Figs 5C1 and 5C4 and S4B1) and mosquitoes supplemented with Enterobacter sp. (Figs 5C3 and 5C6 and S4B3). Conversely, no PM was observed in antibiotic treated individuals (Figs 5C2 and 5C5 and S4B2). These results suggest that the presence of gut microbes is essential to maintain the structural integrity of the PM during blood feeding.
Fig 5. The gut microbiota promotes PM formation.
(A) Culturable gut microbiota density in normal, antibiotic treated (Abx) and antibiotic treated mosquitoes recolonized with 1X105/ml (5), 1X106/ml (6) and 1X107/ml (7) Enterobacter sp. (B) Infection rate of P. berghei in normal, antibiotic treated (Abx) and antibiotic treated mosquitoes recolonized with 1X105/ml (5), 1X106/ml (6) and 1X107/ml (7) of Enterobacter sp. Median oocyst number is indicated by horizontal black bars. Each dot represents an individual mosquito. (C) PM structure was stained by H&E in normal (C1 and C4), antibiotic treated (C2 and C5) and antibiotic treated mosquitoes recolonized with 1X105/ml Enterobacter sp. (C3 and C6) with 100X (C1-3) and 200X (C4-6) magnification. Arrows denote the PM. Images are representative of two independent experiments. Scale bars, 100 μm.
PM influences outcomes of Plasmodium infection
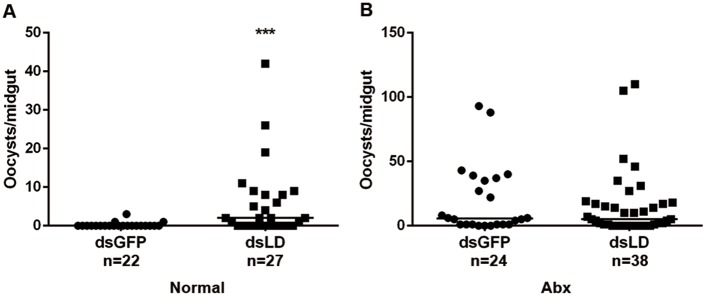
The PM functions as a physical barrier in mosquito that limits Plasmodium infection [31,32]. To further analyze whether the increasing susceptibility in dsLD mosquitoes was due to a compromised PM, we next silenced PGRP-LD in antibiotic treated mosquitoes that lacked a PM and then monitored their susceptibility to parasite infection. In agreement with our previous results, silencing PGRP-LD led to an 8 fold increase in oocyst numbers in dsLD mosquitoes comparing to dsGFP controls (Fig 6A). However, no detectable difference of oocysts number was observed in antibiotic treated mosquitoes injected with dsRNAs (Fig 6B). This result further confirms that enhanced susceptibility to Plasmodium infection in dsLD mosquitoes results from the comprised PM. Together, these results indicate that PGRP-LD helps to maintain homeostasis of the gut microbiota by negatively regulating immune responses. The healthy gut microbes promotes the structural integrity of PM. The intact PM functions as a physical barrier that reduces the capacity of parasites to establish infection in mosquitoes.
Fig 6. Influence of PGRP-LD on parasitism in normal and antibiotic treated mosquitoes.
(A) Oocyst numbers in normal mosquitoes treated with dsRNA. (B) Oocyst numbers in antibiotic treated mosquitoes that received dsRNA. Median oocyst number is indicated by horizontal black bars. Each dot represents an individual mosquito. Significance was determined by Mann-Whitney test. ***, P<0.001. Results from one of three independent experiments are shown.
Discussion
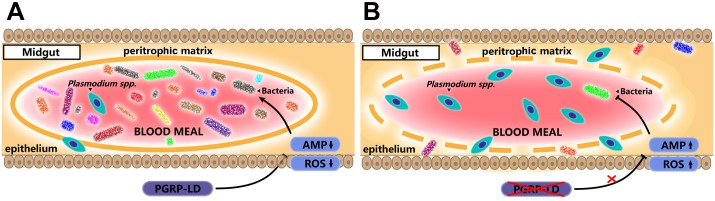
In both invertebrates and vertebrates PGRPs play important roles in regulating interactions with pathogens and commensal bacteria [9]. In this study, we show that PGRP-LD protects A. stephensi from parasite infection by regulating homeostasis of the mosquito’s gut microbiota (Fig 7). Reduced pgrp-ld activates the host immune system, which depletes the abundance of gut microbes in this niche. This impairs PM structure and increases susceptibility to parasite infection.
Fig 7. Model of influence of PGRP-LD on A. stephensi competence.
(A) PGRP-LD protects gut microbiota by negatively regulating immune responses. The gut microbes promote integrity of PM, which enhances vector resistance to parasite infection. (B) Knock down of pgrp-ld leads to the upregulation of immune effectors, which kills a significant number of gut microbes. Dysbiosis of the gut microbiota results in the presentation of a structurally compromised PM, which facilitates P. berghei infection.
PGRP family members were first identified because they share a conserved PGRP domain that is able to detect peptidoglycan (PGN) present on the cell wall of both Gram+ and Gram- bacteria [9,33]. Recent studies using disease vectors show that PGRPs also play important roles in parasite defense [9,12,34–36]. The function of PGRP-LC is well characterized in Anopheles mosquitoes and the tsetse fly, where the protein is responsible for initiating synthesis of downstream effectors in response to both native microbes and invading pathogens [9,11,37]. PGRP-LA participates in defense against parasite infection by functioning similarly to PGRP-LC [12]. PGRP-LB acts as a negative regulator of Imd signaling pathway through its amidase activity [12,38–40]. In tsetse, PGRP-LB has evolved to exhibit bactericidal and anti-parasitic activity [41]. Unlike the above-mentioned PGRPs, little is known about the mechanistic role of PGRP-LD in pathogen defense in either Drosophila or other insects, except that it protects Armigeres mosquitoes from E.coli infection by modulating expression of downstream antimicrobial peptides [42]. We show here that A. stephesi PGRP-LD promotes host defense against P. berghei. Experimental knock down of pgrp-ld expression induces the expression of downstream effectors both in the presence or absence of parasite challenge. Based on its structure, A. stephensi PGRP-LD lacks conserved residues essential for either PGN binding or amidase activity, which has been identified in Drosophila PGRPs [43–45]. This is in contrast with most of PGRPs, which function as negative regulators that prevent over activation of immune signaling pathways by catabolizing immunostimulatory peptidoglycan [46,47]. One explanation is that PGRP-LD may use less well conserved residues to bind peptidoglycan. Alternatively, PGRP-LD may interfere the signal transduction of immune pathways, as does Drosophila PGRP-LF that dampens Imd signaling strength by interfering with PGRP-LC-peptidoglycan binding activity [48]. Further investigations are required to determine how PGRP-LD regulates immune system function.
The gut microbiota enhances host intestinal barrier function and pathogen tolerance in both vertebrates and invertebrates [47,49]. In A. stephensi, pgrp-ld knockdown elevates immune activity that eliminates the majority of gut microbes but fails to eliminate P. berghei. In these mosquitoes, the spatial structure of remaining bacteria was altered. Gut microbes that are usually restrained within the endoperitrophic space localize in close contact with midgut epithelium. Our results indicate that PM structure is compromised in dsLD treated mosquitoes. We then observe PM structure is impaired and expression of PM genes varies significantly. These results suggest that the defect of PM structure results from the dysregulation of PM genes. In addition, we also find that the PM of A. stephensi is absent 48 hr post blood meal in antibiotic treated mosquitoes in which most enteric microbes are cleared. This defect is also associated with decreasing peritrophin and increasing chitinase expression. Thus PM structural integrity is associated with the homeostasis of gut bacteria in A. stephensi, similarly as in many disease vectors [27–29]. When mosquitoes treated with antibiotics are re-colonized by Enterobacter sp, both PM structural integrity and vector competence are restored. In agreement with the finding in An. coluzzii that PM is induced by gut microbiota [29], our results further confirm that gut microbiota of Anopheles mosquitoes is essential for PM integrity. However, we are currently unable to say at what abundance and how gut microbiota are able to maintain intact PM structure.
The PM serves as a physical barrier to parasite infection establishment in multiple disease transmitting vectors, including tsetse flies, sand flies and ticks [27,32,50–56]. Our study shows that in antibiotic treated mosquitoes that present a compromised PM, knockdown of pgrp-ld expression does not change infection prevalence compared to controls. This result reinforces that increasing susceptibility of dsLD mosquitoes to P. berghei infection is due to the comprised PM as opposed to reduced levels as of PGRP-LD directly. In agreement with most vectors, our results show that PM is a major physical barrier that prevents P. berghei infection establishment in A. stephensi.
In summary, our data demonstrate that a complex interplay exists between the host immune system, gut microbes and the PM, and this interplay determines parasite infection outcomes in A. stephensi. PGRP-LD, functioning as a key mediator, helps to maintain this balance. Detailed studies on the regulation of PGRP-LD on immune signaling pathways, and the influence of gut microbiota on PM formation, are currently under way and may provide new insights into interactions between immune system, gut microbiota and parasites.
Materials and methods
Ethics statement
All animals were handled according to the guidelines for the Care and Use of Laboratory Animals of the National Institutes of Health and the Office of Laboratory Animal Welfare. The research protocol was approved by the Institutional animal care and use committee, Department of Laboratory Animal Science, Fudan University (IACUC 20161784A359).
Mosquito rearing and antibiotic treatment
The Anopheles stephensi mosquito (strain Hor) was reared at 28°C, 80% relative humidity and at a 12h light/dark cycle. Adults were maintained on 10% sucrose and BALB/c mice. Newly eclosed mosquitoes were administrated with fresh filtered 10% sucrose supplemented with 10 U/ml penicillin, 10 μg/ml streptomycin and 15 μg/ml gentamicin daily, for up to 5 days [13].
Gene silencing and reverse transcription quantitative PCR (RT-qPCR)
PCR amplicons tailed with T7 promoter sequences were used to synthesize dsRNAs using MEGAscript RNA kit (Ambion, Invitrogen). The cDNA clones Astepgrp-ld (ASTE010245), and plasmid eGFP (BD Biosciences) served as templates for amplification using gene specific primers (S1 Table). Five to 6-day-old females received a total 69 nl dsRNAs (4μg/μl) injected intra-thoracically using nanoject II microinjector (Drummond). Injected mosquitoes were allowed to recover for 5 days prior to infection [57]. Survival rate was recorded daily for 5 days post dsRNA treatments and compared to that of dsGFP controls. Silencing efficiency was verified by qPCR 2-day post dsRNA treatment with primers listed in S1 Table. RNA was extracted from flash frozen mosquitoes utilizing the standard TRI reagent (Sigma-Aldrich, China) protocol. cDNA was prepared from total RNA using the 5XAll-in-One MasterMix (with AccuRT Genomic DNA Removal Kit) (ABM, China). Levels of target genes were determined by Roche LightCycler 96 Real Time PCR Detection System with SYBR Green qPCR Master Mix (Biomake, China) using the following conditions: 95°C for 5 min, 40 cycles of 15 sec at 95°C, 30 sec at 60°C, and 15 sec at 72°C. Fluorescence readings were taken at 72°C after each cycle. Melting curves (60°C–95°C) were performed to confirm the identity of the PCR product. The data were processed and analyzed with LightCycler 96 software. Expression of cecropin, gambicin, defensin, tep1, prophenoloxidase, nos, duox and caudal were analyzed 5 days post dsRNA administration with primers listed in S1 Table. Ribosomal gene S7 widely used in studies of Anopheles gene expression was used as the internal reference [58–61]. PCR efficiency of each primer set was determined by standard curve. Relative quantitation results were normalized with S7 and analyzed by the 2–ΔΔCt method [62]. Gene expression of dsLD treated group was normalized to dsGFP controls. The normality of data sets was determined by Shapiro-Wilk test before t test analysis. Values are represented as the mean (±SEM), and statistical significance was determined using a Student’s t test and Excel software.
Plasmodium infection
A. stephensi were starved overnight and then fed on P. berghei (ANKA) infected BALB/c with parasitemia of 6–7% using standard protocols [63]. Mosquitoes were starved for 24 hr before blood feeding. After imbibing a blood meal, mosquitoes were maintained at 21°C. Un-engorged mosquitoes were removed 24hr post blood meal. Midguts were dissected and infection intensity were determined microscopically 8-day post infection. The oocyst data were not normally distributed as determined by Shapiro-Wilk test. Thus, significance was determined using the Mann-Whitney test.
Microbiota analysis
Mosquitoes were collected 5 day after dsRNA treatment or antibiotic treatment and surface sterilized with 70% ethanol twice and 0.9% NaCl twice. Midguts were dissected and homogenized in 0.9% NaCl. Homogenates were serially diluted and plated on LB agar plates. CFUs were counted 2 days after incubation at 28°C. Total DNA was extracted by the method of Holmes and Bonner as described [64]. Bacterial density was quantified by qPCR using universal 16S rRNA primers [28] (S1 Table). Ribosomal gene S7 was used as the internal reference. Significance was determined using the Student’s t-test.
The composition of the gut microbiota in dsRNA treated mosquitoes was analyzed by pyrosequencing that targeted the V3-V4 region of bacterial 16S rRNA [65]. 10 midguts of dsRNA treated mosquitoes were pooled for 1 biological replicate. DNA of 3 biological replicates of each treatment were prepared for further sequencing analysis (S1 Text).
For fluorescent in situ hybridization (FISH), abdomens of dsLD treated females 2 day post blood meal were fixed and sectioned as described [66]. Slides were hybridized with 10ng/μl universal 16S ribosomal RNA probe (5’-GCTGCCTCCCGTAGGAGT-3’) labeled with Alexa Fluor 555 (Life technology). Tissues were visualized using Nikon ECLIPSE IVi microscope connected to a Nikon DIGITAL SIGHT DS-U3 digital camera.
Peritrophic matrix analysis
Forty-eight hour post blood meal mosquito abdomens were fixed and sectioned as described above [66]. Samples were sectioned at 5 μm, stained with hematoxylin and eosin (H&E) (Huntz Enterprises Inc., China) and Periodic Acid Schiff (PAS) (Sigma-Aldrich, China) according to the manufacturer’s protocol. Slides were hard mounted using Canada balsam (ChemsWorth). Slides were viewed using bright field illumination on a Nikon ECLIPSE IVi microscope connected to a Nikon DIGITAL SIGHT DS-U3 digital camera. Four days post dsRNA treatment A. stephensi were fed with blood meal supplemented with 500 kDa FITC-labeled dextran molecules (2.5mg/ml blood)(Sigma) which were filtered using PD MiniTrap Sephadex G10 columns (GE Healthcare) as described [27]. Forty eight hours post-feeding, midguts were dissected and FITC signal observed using a Zeiss, LSM710 confocal microscope connected to a Nikon DIGITAL SIGHT DS-U3 digital camera. Expression of 4 PM genes was analyzed 24 hr and 48 hr post blood meal using primers targeting peritrophin1(ASTE010406), peritrophin14 (ASTE009456), 2 chitinases, herein named chitinaseA (ASTE005630) and chitinaseB (ASTE000328) (S1 Table).
Oral administration of bacteria
The administration of mosquito commensal bacteria was performed as described [67]. Briefly, an overnight culture of Enterobacter sp. was washed 2 times in phosphate-buffered saline (PBS) and introduced to mosquitoes via a sugar meal. A final concentration of 1X105~1X107 /ml bacteria was added to 1.5% sterile sugar. All mosquitoes were starved for 24hr hours before being offered a blood meal. Mosquitoes were given a blood meal 2-day post bacteria treatment. Age matched wild-type and antibiotic treated mosquitoes were used as controls.
Supporting information
(DOCX)
(DOCX)
PGRPs from Anopheles stephensi: AstePGRP-LD (ASTE010245), AstePGRP-LB (ASTE006009); Anopheles gambiae: AgPGRP-LC (AGAP005203), AgPGRP-LD (AGAP005552), AgGPRP-LB (AGAP001212); Drosophila melanogaster: DmPGRP-LCx (FBGN0035976), DmPGRP-LB (FBGN0037906), DmPGRP-LD (FBGN0260458), DmPGRP-SA (FBGN0030310) and Glossina morsitans morsitans: GmmPGRP-LC (GMOY006094), GmmPGRP-LD (GMOY004195), GmmPGRP-LB (GMOY006730). Three conserved PGRP domains are boxed in black and numbered. The highly conserved residues among all PGRP proteins are shown in grey, conserved residues present in the recognition PGRPs and catalytic PGRPs are shown in light grey shadow. Residues required for amidase activity are indicated by a star at the bottom. Residues required for peptidoglycan binding in DmPGRP-LCx, DmPGRP-SA are indicated by diamond and triangle, respectively.
(TIF)
Survival was recorded daily for 5 days post dsRNA treatments and compared to that of dsGFP controls. No significant difference was seen between dsLD and dsGFP mosquitoes. The data are the representative of three replicate infections. Total sample size: dsGFP (n = 30), dsLD (n = 30).
(TIF)
(A) Incidence of the major bacterial taxonomic in dsGFP and dsLD. Relative abundance of identified microbial taxa in the midguts collected from mosquitoes 5 day post dsRNA treatment. (B) Principal coordinates analysis of the bacterial composition in dsGFP (red dots) and dsLD (green dots) at operational taxonomic unit (OTU) (97%) level. Each sample contains 10 midguts.
(TIF)
(A) PM structure was observed in dsGFP (A1) and dsLD (A2) mosquitoes at 200X magnification. (B) PM structure was observed in normal (B1), antibiotic treated mosquitoes (B2) and antibiotic treated mosquitoes recolonized with 1X105/ml Enterobacter sp. (B3) at 200X magnification. Arrows denote the PM. Images are representative of three independent experiments. Scale bars, 50 μm.
(TIF)
Error bars indicate standard error. Images are representative of three independent experiments.
(TIF)
Acknowledgments
We thank Dr. Bin Xu from National Institute of Parasitic Diseases for providing Plasmodium berghei ANKA strain. We thank Dr. Brian Weiss (Yale School of Public Health) for critical review of the manuscript.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This research was supported by National Research and Development Plan of China (No. 2016YFC1200500) (http://program.most.gov.cn/), National Natural Science Foundation of China (31472039) (https://isisn.nsfc.gov.cn/egrantweb/), National Institutes of Health Grant (R01AI129819) (https://grants.nih.gov/grants) and the Research Fund of the State Key Laboratory of Genetic Engineering, Fudan University. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.World Health Organization (2015) World Malaria Report 2015.
- 2.Wang S, Jacobs-Lorena M (2013) Genetic approaches to interfere with malaria transmission by vector mosquitoes. Trends Biotechnol 31: 185–193. doi: 10.1016/j.tibtech.2013.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clayton AM, Dong Y, Dimopoulos G (2014) The Anopheles innate immune system in the defense against malaria infection. J Innate Immun 6: 169–181. doi: 10.1159/000353602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marois E (2011) The multifaceted mosquito anti-Plasmodium response. Curr Opin Microbiol 14: 429–435. doi: 10.1016/j.mib.2011.07.016 [DOI] [PubMed] [Google Scholar]
- 5.Hegedus D, Erlandson M, Gillott C, Toprak U (2009) New insights into peritrophic matrix synthesis, architecture, and function. Annu Rev Entomol 54: 285–302. doi: 10.1146/annurev.ento.54.110807.090559 [DOI] [PubMed] [Google Scholar]
- 6.Han YS, Thompson J, Kafatos FC, Barillas-Mury C (2000) Molecular interactions between Anopheles stephensi midgut cells and Plasmodium berghei: the time bomb theory of ookinete invasion of mosquitoes. EMBO J 19: 6030–6040. doi: 10.1093/emboj/19.22.6030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kakani P, Suman S, Gupta L, Kumar S (2016) Ambivalent Outcomes of Cell Apoptosis: A Barrier or Blessing in Malaria Progression. Front Microbiol 7: 302 doi: 10.3389/fmicb.2016.00302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rodrigues J, Brayner FA, Alves LC, Dixit R, Barillas-Mury C (2010) Hemocyte differentiation mediates innate immune memory in Anopheles gambiae mosquitoes. Science 329: 1353–1355. doi: 10.1126/science.1190689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Royet J, Gupta D, Dziarski R (2011) Peptidoglycan recognition proteins: modulators of the microbiome and inflammation. Nat Rev Immunol 11: 837–851. doi: 10.1038/nri3089 [DOI] [PubMed] [Google Scholar]
- 10.Christophides GK, Zdobnov E, Barillas-Mury C, Birney E, Blandin S, et al. (2002) Immunity-related genes and gene families in Anopheles gambiae. Science 298: 159–165. doi: 10.1126/science.1077136 [DOI] [PubMed] [Google Scholar]
- 11.Meister S, Agianian B, Turlure F, Relogio A, Morlais I, et al. (2009) Anopheles gambiae PGRPLC-mediated defense against bacteria modulates infections with malaria parasites. PLoS Pathog 5: e1000542 doi: 10.1371/journal.ppat.1000542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gendrin M, Turlure F, Rodgers FH, Cohuet A, Morlais I, et al. (2017) The Peptidoglycan Recognition Proteins PGRPLA and PGRPLB Regulate Anopheles Immunity to Bacteria and Affect Infection by Plasmodium. J Innate Immun 9: 333–342. doi: 10.1159/000452797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dong Y, Manfredini F, Dimopoulos G (2009) Implication of the mosquito midgut microbiota in the defense against malaria parasites. PLoS Pathog 5: e1000423 doi: 10.1371/journal.ppat.1000423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ricci I, Damiani C, Capone A, DeFreece C, Rossi P, et al. (2012) Mosquito/microbiota interactions: from complex relationships to biotechnological perspectives. Curr Opin Microbiol 15: 278–284. doi: 10.1016/j.mib.2012.03.004 [DOI] [PubMed] [Google Scholar]
- 15.Ramirez JL, Short SM, Bahia AC, Saraiva RG, Dong Y, et al. (2014) Chromobacterium Csp_P reduces malaria and dengue infection in vector mosquitoes and has entomopathogenic and in vitro anti-pathogen activities. PLoS Pathog 10: e1004398 doi: 10.1371/journal.ppat.1004398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xiao X, Yang L, Pang X, Zhang R, Zhu Y, et al. (2017) A Mesh-Duox pathway regulates homeostasis in the insect gut. Nat Microbiol 2: 17020 doi: 10.1038/nmicrobiol.2017.20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pang X, Xiao X, Liu Y, Zhang R, Liu J, et al. (2016) Mosquito C-type lectins maintain gut microbiome homeostasis. Nat Microbiol 1: 16023. [DOI] [PubMed] [Google Scholar]
- 18.Cirimotich CM, Dong Y, Clayton AM, Sandiford SL, Souza-Neto JA, et al. (2011) Natural microbe-mediated refractoriness to Plasmodium infection in Anopheles gambiae. Science 332: 855–858. doi: 10.1126/science.1201618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clayton AM, Cirimotich CM, Dong Y, Dimopoulos G (2013) Caudal is a negative regulator of the Anopheles IMD pathway that controls resistance to Plasmodium falciparum infection. Dev Comp Immunol 39: 323–332. doi: 10.1016/j.dci.2012.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yassine H, Osta MA (2010) Anopheles gambiae innate immunity. Cell Microbiol 12: 1–9. doi: 10.1111/j.1462-5822.2009.01388.x [DOI] [PubMed] [Google Scholar]
- 21.Oliveira JH, Goncalves RL, Lara FA, Dias FA, Gandara AC, et al. (2011) Blood meal-derived heme decreases ROS levels in the midgut of Aedes aegypti and allows proliferation of intestinal microbiota. PLoS Pathog 7: e1001320 doi: 10.1371/journal.ppat.1001320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gusmao DS, Santos AV, Marini DC, Bacci M Jr., Berbert-Molina MA, et al. (2010) Culture-dependent and culture-independent characterization of microorganisms associated with Aedes aegypti (Diptera: Culicidae) (L.) and dynamics of bacterial colonization in the midgut. Acta Trop 115: 275–281. doi: 10.1016/j.actatropica.2010.04.011 [DOI] [PubMed] [Google Scholar]
- 23.Lehane MJ (1997) Peritrophic matrix structure and function. Annu Rev Entomol 42: 525–550. doi: 10.1146/annurev.ento.42.1.525 [DOI] [PubMed] [Google Scholar]
- 24.Engel P, Moran NA (2013) The gut microbiota of insects—diversity in structure and function. FEMS Microbiol Rev 37: 699–735. doi: 10.1111/1574-6976.12025 [DOI] [PubMed] [Google Scholar]
- 25.Dinglasan RR, Devenport M, Florens L, Johnson JR, McHugh CA, et al. (2009) The Anopheles gambiae adult midgut peritrophic matrix proteome. Insect Biochem Mol Biol 39: 125–134. doi: 10.1016/j.ibmb.2008.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhu Q, Arakane Y, Banerjee D, Beeman RW, Kramer KJ, et al. (2008) Domain organization and phylogenetic analysis of the chitinase-like family of proteins in three species of insects. Insect Biochem Mol Biol 38: 452–466. doi: 10.1016/j.ibmb.2007.06.010 [DOI] [PubMed] [Google Scholar]
- 27.Weiss BL, Wang J, Maltz MA, Wu Y, Aksoy S (2013) Trypanosome infection establishment in the tsetse fly gut is influenced by microbiome-regulated host immune barriers. PLoS Pathog 9: e1003318 doi: 10.1371/journal.ppat.1003318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Narasimhan S, Rajeevan N, Liu L, Zhao YO, Heisig J, et al. (2014) Gut microbiota of the tick vector Ixodes scapularis modulate colonization of the Lyme disease spirochete. Cell Host Microbe 15: 58–71. doi: 10.1016/j.chom.2013.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rodgers FH, Gendrin M, Wyer CAS, Christophides GK (2017) Microbiota-induced peritrophic matrix regulates midgut homeostasis and prevents systemic infection of malaria vector mosquitoes. PLoS Pathog 13: e1006391 doi: 10.1371/journal.ppat.1006391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eappen AG, Smith RC, Jacobs-Lorena M (2013) Enterobacter-activated mosquito immune responses to Plasmodium involve activation of SRPN6 in Anopheles stephensi. PLoS One 8: e62937 doi: 10.1371/journal.pone.0062937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Abraham EG, Jacobs-Lorena M (2004) Mosquito midgut barriers to malaria parasite development. Insect Biochem Mol Biol 34: 667–671. doi: 10.1016/j.ibmb.2004.03.019 [DOI] [PubMed] [Google Scholar]
- 32.Billingsley PF, Rudin W (1992) The role of the mosquito peritrophic membrane in bloodmeal digestion and infectivity of Plasmodium species. J Parasitol 78: 430–440. [PubMed] [Google Scholar]
- 33.Dziarski R, Gupta D (2010) Review: Mammalian peptidoglycan recognition proteins (PGRPs) in innate immunity. Innate Immun 16: 168–174. doi: 10.1177/1753425910366059 [DOI] [PubMed] [Google Scholar]
- 34.Yano T, Kurata S (2011) Intracellular recognition of pathogens and autophagy as an innate immune host defence. J Biochem 150: 143–149. doi: 10.1093/jb/mvr083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang S, Beerntsen BT (2015) Functional implications of the peptidoglycan recognition proteins in the immunity of the yellow fever mosquito, Aedes aegypti. Insect Mol Biol 24: 293–310. doi: 10.1111/imb.12159 [DOI] [PubMed] [Google Scholar]
- 36.Wang S, Beerntsen BT (2013) Insights into the different functions of multiple peptidoglycan recognition proteins in the immune response against bacteria in the mosquito, Armigeres subalbatus. Insect Biochem Mol Biol 43: 533–543. doi: 10.1016/j.ibmb.2013.03.004 [DOI] [PubMed] [Google Scholar]
- 37.Wang J, Wu Y, Yang G, Aksoy S (2009) Interactions between mutualist Wigglesworthia and tsetse peptidoglycan recognition protein (PGRP-LB) influence trypanosome transmission. Proc Natl Acad Sci U S A 106: 12133–12138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Anselme C, Perez-Brocal V, Vallier A, Vincent-Monegat C, Charif D, et al. (2008) Identification of the weevil immune genes and their expression in the bacteriome tissue. BMC Biol 6: 43 doi: 10.1186/1741-7007-6-43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zaidman-Remy A, Herve M, Poidevin M, Pili-Floury S, Kim MS, et al. (2006) The Drosophila amidase PGRP-LB modulates the immune response to bacterial infection. Immunity 24: 463–473. doi: 10.1016/j.immuni.2006.02.012 [DOI] [PubMed] [Google Scholar]
- 40.Ratzka C, Gross R, Feldhaar H (2013) Gene expression analysis of the endosymbiont-bearing midgut tissue during ontogeny of the carpenter ant Camponotus floridanus. J Insect Physiol 59: 611–623. doi: 10.1016/j.jinsphys.2013.03.011 [DOI] [PubMed] [Google Scholar]
- 41.Wang J, Aksoy S (2012) PGRP-LB is a maternally transmitted immune milk protein that influences symbiosis and parasitism in tsetse’s offspring. Proc Natl Acad Sci U S A 109: 10552–10557. doi: 10.1073/pnas.1116431109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang S, Beerntsen BT (2014) Identification and functional analysis of the peptidoglycan recognition protein LD gene in the mosquito, Armigeres subalbatus. Dev Comp Immunol 42: 148–158. doi: 10.1016/j.dci.2013.08.025 [DOI] [PubMed] [Google Scholar]
- 43.Chang CI, Chelliah Y, Borek D, Mengin-Lecreulx D, Deisenhofer J (2006) Structure of tracheal cytotoxin in complex with a heterodimeric pattern-recognition receptor. Science 311: 1761–1764. doi: 10.1126/science.1123056 [DOI] [PubMed] [Google Scholar]
- 44.Chang CI, Pili-Floury S, Herve M, Parquet C, Chelliah Y, et al. (2004) A Drosophila pattern recognition receptor contains a peptidoglycan docking groove and unusual L,D-carboxypeptidase activity. PLoS Biol 2: E277 doi: 10.1371/journal.pbio.0020277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim MS, Byun M, Oh BH (2003) Crystal structure of peptidoglycan recognition protein LB from Drosophila melanogaster. Nat Immunol 4: 787–793. doi: 10.1038/ni952 [DOI] [PubMed] [Google Scholar]
- 46.Paredes JC, Welchman DP, Poidevin M, Lemaitre B (2011) Negative regulation by amidase PGRPs shapes the Drosophila antibacterial response and protects the fly from innocuous infection. Immunity 35: 770–779. doi: 10.1016/j.immuni.2011.09.018 [DOI] [PubMed] [Google Scholar]
- 47.Buchon N, Broderick NA, Lemaitre B (2013) Gut homeostasis in a microbial world: insights from Drosophila melanogaster. Nat Rev Microbiol 11: 615–626. doi: 10.1038/nrmicro3074 [DOI] [PubMed] [Google Scholar]
- 48.Maillet F, Bischoff V, Vignal C, Hoffmann J, Royet J (2008) The Drosophila peptidoglycan recognition protein PGRP-LF blocks PGRP-LC and IMD/JNK pathway activation. Cell Host Microbe 3: 293–303. doi: 10.1016/j.chom.2008.04.002 [DOI] [PubMed] [Google Scholar]
- 49.Bron PA, Kleerebezem M, Brummer RJ, Cani PD, Mercenier A, et al. (2017) Can probiotics modulate human disease by impacting intestinal barrier function? Br J Nutr 117: 93–107. doi: 10.1017/S0007114516004037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Boucias D, Baniszewski J, Prompiboon P, Lietze V, Geden C (2015) Enhancement of the Musca domestica hytrosavirus infection with orally delivered reducing agents. J Invertebr Pathol 124: 35–43. doi: 10.1016/j.jip.2014.10.004 [DOI] [PubMed] [Google Scholar]
- 51.Weiss BL, Savage AF, Griffith BC, Wu Y, Aksoy S (2014) The peritrophic matrix mediates differential infection outcomes in the tsetse fly gut following challenge with commensal, pathogenic, and parasitic microbes. J Immunol 193: 773–782. doi: 10.4049/jimmunol.1400163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Coutinho-Abreu IV, Sharma NK, Robles-Murguia M, Ramalho-Ortigao M (2013) Characterization of Phlebotomus papatasi peritrophins, and the role of PpPer1 in Leishmania major survival in its natural vector. PLoS Negl Trop Dis 7: e2132 doi: 10.1371/journal.pntd.0002132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shahabuddin M, Toyoshima T, Aikawa M, Kaslow DC (1993) Transmission-blocking activity of a chitinase inhibitor and activation of malarial parasite chitinase by mosquito protease. Proc Natl Acad Sci U S A 90: 4266–4270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vinetz JM, Valenzuela JG, Specht CA, Aravind L, Langer RC, et al. (2000) Chitinases of the avian malaria parasite Plasmodium gallinaceum, a class of enzymes necessary for parasite invasion of the mosquito midgut. J Biol Chem 275: 10331–10341. [DOI] [PubMed] [Google Scholar]
- 55.Kato N, Mueller CR, Fuchs JF, McElroy K, Wessely V, et al. (2008) Evaluation of the function of a type I peritrophic matrix as a physical barrier for midgut epithelium invasion by mosquito-borne pathogens in Aedes aegypti. Vector Borne Zoonotic Dis 8: 701–712. doi: 10.1089/vbz.2007.0270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Abraham NM, Liu L, Jutras BL, Yadav AK, Narasimhan S, et al. (2017) Pathogen-mediated manipulation of arthropod microbiota to promote infection. Proc Natl Acad Sci U S A 114: E781–E790. doi: 10.1073/pnas.1613422114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Blandin S, Moita LF, Kocher T, Wilm M, Kafatos FC, et al. (2002) Reverse genetics in the mosquito Anopheles gambiae: targeted disruption of the Defensin gene. EMBO Rep 3: 852–856. doi: 10.1093/embo-reports/kvf180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dimopoulos G, Richman A, della Torre A, Kafatos FC, Louis C (1996) Identification and characterization of differentially expressed cDNAs of the vector mosquito, Anopheles gambiae. Proc Natl Acad Sci U S A 93: 13066–13071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dimopoulos G, Richman A, Muller HM, Kafatos FC (1997) Molecular immune responses of the mosquito Anopheles gambiae to bacteria and malaria parasites. Proc Natl Acad Sci U S A 94: 11508–11513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gupta L, Molina-Cruz A, Kumar S, Rodrigues J, Dixit R, et al. (2009) The STAT pathway mediates late-phase immunity against Plasmodium in the mosquito Anopheles gambiae. Cell Host Microbe 5: 498–507. doi: 10.1016/j.chom.2009.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kumar S, Molina-Cruz A, Gupta L, Rodrigues J, Barillas-Mury C (2010) A peroxidase/dual oxidase system modulates midgut epithelial immunity in Anopheles gambiae. Science 327: 1644–1648. doi: 10.1126/science.1184008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25: 402–408. doi: 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 63.Sinden RE (1997) Infection of mosquitoes with rodent malaria In: Crampton JM, Beard C.B., and Christos L., editor. The Molecular Biology of Insect Disease Vectors: Netherlands: Springer; pp. 67–91. [Google Scholar]
- 64.Holmes DS, Bonner J (1973) Preparation, molecular weight, base composition, and secondary structure of giant nuclear ribonucleic acid. Biochemistry 12: 2330–2338. [DOI] [PubMed] [Google Scholar]
- 65.Michelsen CF P P, Glaring MA, Schjoerring JK, Stougaard P (2014) Bacterial diversity in Greenlandic soils as affected by potato cropping and inorganic versus organic fertilization. Polar Biology 37: 61–71. [Google Scholar]
- 66.Attardo GM, Lohs C, Heddi A, Alam UH, Yildirim S, et al. (2008) Analysis of milk gland structure and function in Glossina morsitans: milk protein production, symbiont populations and fecundity. J Insect Physiol 54: 1236–1242. doi: 10.1016/j.jinsphys.2008.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bahia AC, Dong Y, Blumberg BJ, Mlambo G, Tripathi A, et al. (2014) Exploring Anopheles gut bacteria for Plasmodium blocking activity. Environ Microbiol 16: 2980–2994. doi: 10.1111/1462-2920.12381 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(DOCX)
PGRPs from Anopheles stephensi: AstePGRP-LD (ASTE010245), AstePGRP-LB (ASTE006009); Anopheles gambiae: AgPGRP-LC (AGAP005203), AgPGRP-LD (AGAP005552), AgGPRP-LB (AGAP001212); Drosophila melanogaster: DmPGRP-LCx (FBGN0035976), DmPGRP-LB (FBGN0037906), DmPGRP-LD (FBGN0260458), DmPGRP-SA (FBGN0030310) and Glossina morsitans morsitans: GmmPGRP-LC (GMOY006094), GmmPGRP-LD (GMOY004195), GmmPGRP-LB (GMOY006730). Three conserved PGRP domains are boxed in black and numbered. The highly conserved residues among all PGRP proteins are shown in grey, conserved residues present in the recognition PGRPs and catalytic PGRPs are shown in light grey shadow. Residues required for amidase activity are indicated by a star at the bottom. Residues required for peptidoglycan binding in DmPGRP-LCx, DmPGRP-SA are indicated by diamond and triangle, respectively.
(TIF)
Survival was recorded daily for 5 days post dsRNA treatments and compared to that of dsGFP controls. No significant difference was seen between dsLD and dsGFP mosquitoes. The data are the representative of three replicate infections. Total sample size: dsGFP (n = 30), dsLD (n = 30).
(TIF)
(A) Incidence of the major bacterial taxonomic in dsGFP and dsLD. Relative abundance of identified microbial taxa in the midguts collected from mosquitoes 5 day post dsRNA treatment. (B) Principal coordinates analysis of the bacterial composition in dsGFP (red dots) and dsLD (green dots) at operational taxonomic unit (OTU) (97%) level. Each sample contains 10 midguts.
(TIF)
(A) PM structure was observed in dsGFP (A1) and dsLD (A2) mosquitoes at 200X magnification. (B) PM structure was observed in normal (B1), antibiotic treated mosquitoes (B2) and antibiotic treated mosquitoes recolonized with 1X105/ml Enterobacter sp. (B3) at 200X magnification. Arrows denote the PM. Images are representative of three independent experiments. Scale bars, 50 μm.
(TIF)
Error bars indicate standard error. Images are representative of three independent experiments.
(TIF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.