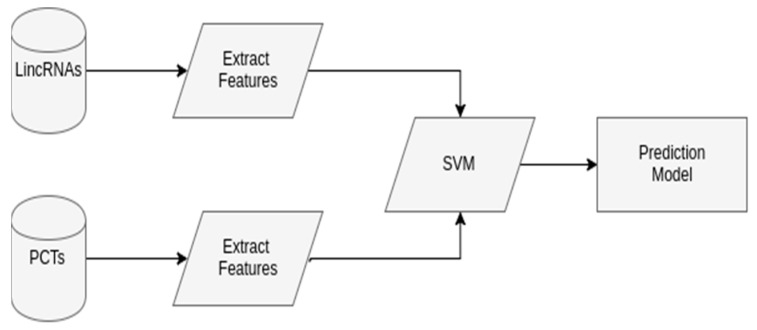
Figure 4.
The generic support vector machine (SVM) model to predict long intergenic non-coding RNA (lincRNAs) in plants, with two phases. In Extract features, a given transcript is screened, so that features that characterize the lincRNAs and protein coding transcripts (PCTs) are extracted, in this case, open reading frames (ORF) length, ORF proportion (ORF length divided by the transcript length), and the 10 more significant 2-, 3-, and 4-nucleotides to identify lincRNAs according to principal component analysis (PCA). In SVM, the SVM model is constructed, with balanced data of lincRNAs and PCTs features, 10-fold cross-validation, grid-search, and using 80% of the data as the training set and the other 20% as test. The input data include lincRNAs as the positive set, and PCTs as the negative set. The output is the constructed SVM model to predict lincRNAs.