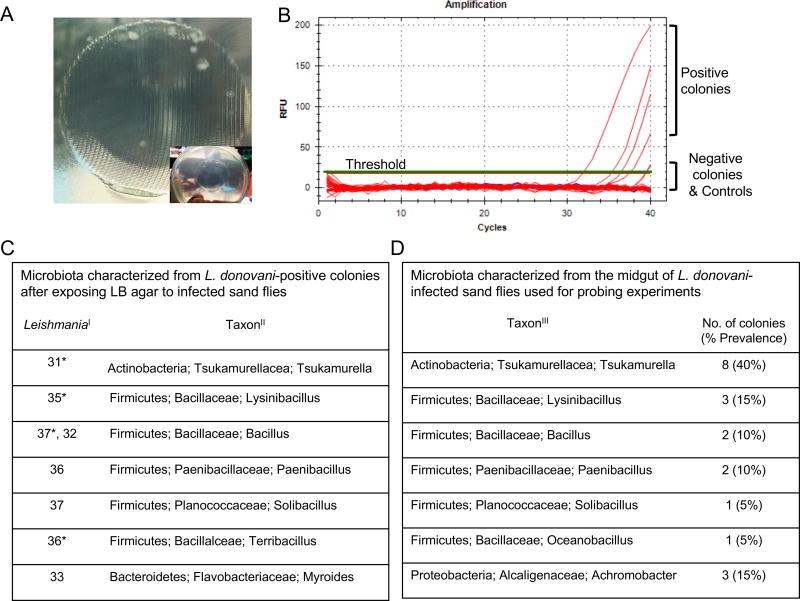
Figure 3. Identification of gut microbes egested during infected sand fly bites.
(A) Bacterial colonies growing on LB/Agar after exposure to L. donovani-infected sand flies. Inset shows the mesh imprint of the feeder containing the sand flies. Pictures representative of five independent experiments.
(B) Representative Real-Time PCR amplification curves (red lines) for Leishmania minicircle kDNA using bacterial colonies picked from agar plates as template. Green line, Threshold. Blue Lines, Negative controls (water or agar).
(C) Bacteria identified from Leishmania-positive colonies. I, Real-time PCR CT values for amplified Leishmania. II, Taxonomy of bacteria ordered by phylum, family, and genus. *, colonies depicted in (B).
(D) Bacteria identified from the midgut of infected sand fly groups used in (A). III, Taxonomy of bacteria ordered by phylum, family, and genus.
See also Video S1.