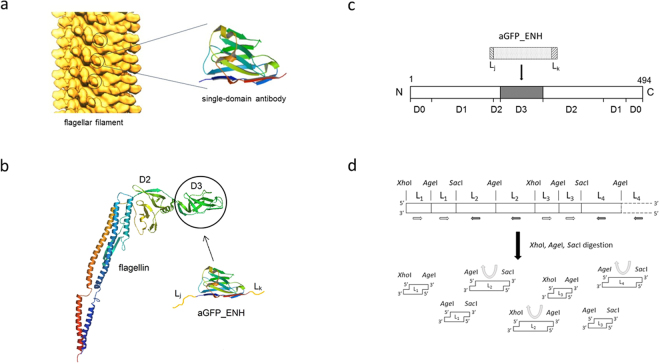
Figure 1.
Construction of flagellar nanotubes displaying single-domain antibodies. (a) Arrangement of flagellin subunits within the flagellar filament. The outer hypervariable D3 portions (circled) of flagellin subunits, situated on the filament surface, were replaced by single-domain antibodies. The distance between the solvent-exposed nanobodies on the filament surface is about 5.5 nm. Solid surface representation of a longitudinal section of the filament according to Mimori-Kiyosue et al. 35. (Copyright (1996) National Academy of Sciences, USA.) (b) The Ca backbone trace of flagellin (PDB code: 1UCU) and aGFP_ENH nanobody (PDB code: 3K1K). The aGFP_ENH nanobody with various oligopeptide linkers attached to both ends was inserted into the middle part of flagellin to replace the D3 domain. (c) Representation of the domain arrangement of flagellin. D0-D2 are discontinuous domains formed by segments from both the N- and C-termini. D3 is constructed by the middle portion of the polypeptide chain consisting of residues 190–283. This internal segment was removed and replaced by a single-domain antibody. (d) Oligonucleotide segments coding for the chosen oligopeptide linkers (L1, L2, …) were joined via segments encoding the cleavage sites of XhoI, SacI or AgeI restriction enzymes to form a multilinker gene. Arrows show the local direction of the reading frame. In the regions indicated by white and black arrows linker segments are coded by the opposite strands of DNA. The multilinker gene was synthesized and fragmented by a mixture of restriction enzymes (XhoI, SacI, AgeI) to obtain linker-encoding segments (shown in the same orientation relative to the reading frame) with sticky ends facilitating construction of the FliC-aGFP_ENH fusion genes (see text).