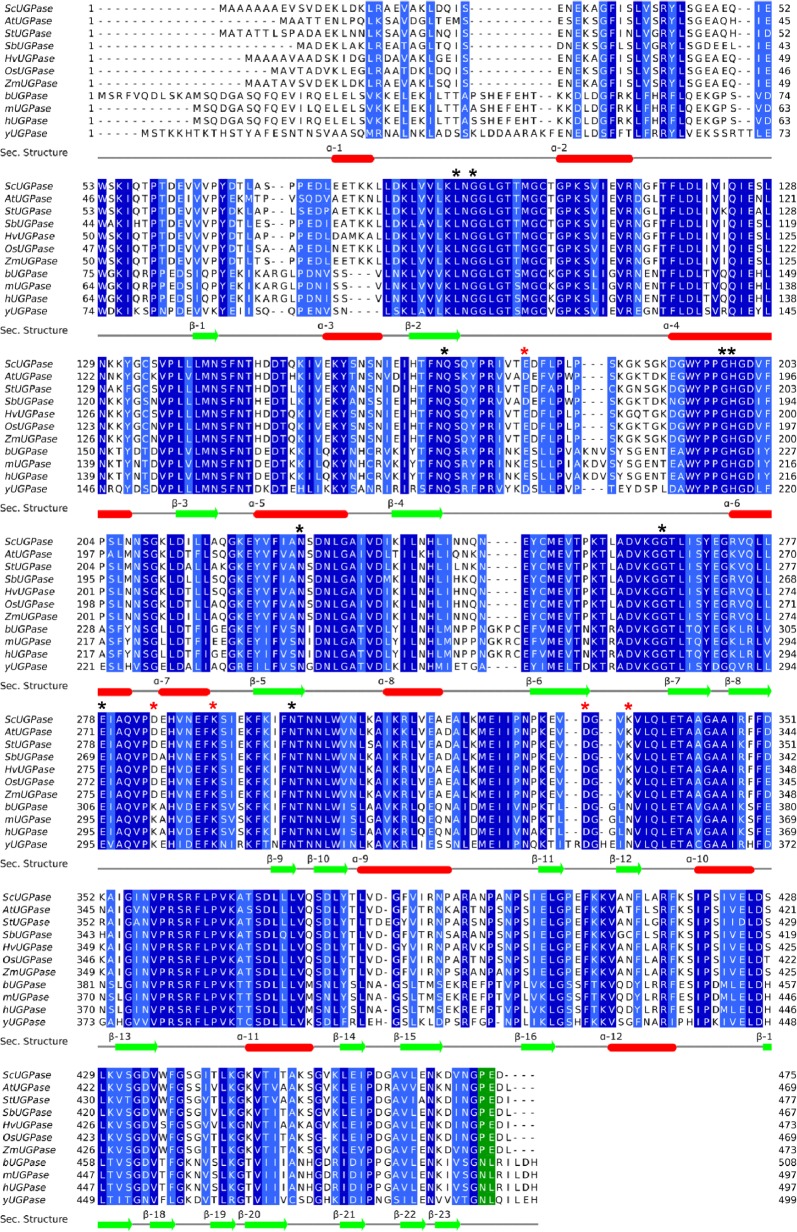
Fig 3. Multiple sequence alignment of UGPase orthologs.
Proteins were aligned by MUSCLE [21] and the alignment optimized in Jalview [22]. The aligned sequences from top to bottom with their accession numbers are: ScUGPase-1 from Saccharum spp—sugarcane (A0A075E2Q1); AtUGPase from Arabidopsis thaliana (Q9M9P3); StUGPase from Solanum tuberosum–potato (P19595); SbUGPase from Sorghum bicolor (C5XSC5); HvUGPase from Hordeum vulgare–barley (Q43772); OsUGPase from Oryza sativa–rice (Q93X08); ZmUGPase from Zea mays–maize (B6T4R3); bUGPase from bovine–Bos taurus (Q07130); mUGPase from mouse–Mus musculus (Q91ZJ5-2); hUGPase from human–Homo sapiens (Q16851-2) and yUGPase from yeast–Saccharomyces cerevisiae (P32861). Region coloured in green corresponds to the residues in the C-terminal region involved in the end-to-end interactions in the yeast and human orthologs. Black asterisks (*) indicate the residues important for ligand binding, and red asterisks (*) indicate residues involved in the dimer interface in the crystal of ScUGPase-1. Elements of secondary structure are shown based on the crystal structure of ScUGPase-1. β-sheets and α-helices are shown in green and red, respectively.