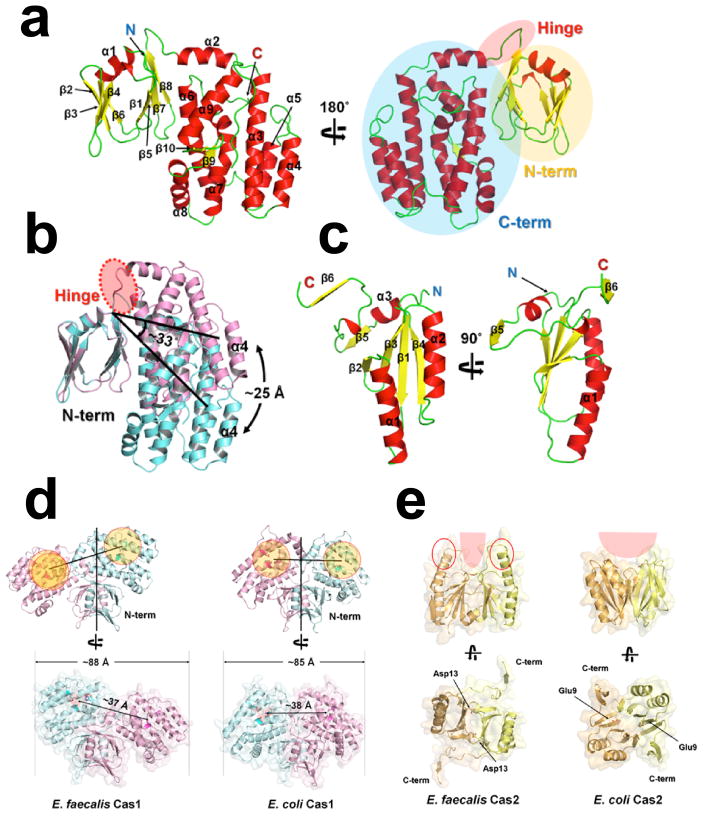
Extended Data Figure 2. Comparison of the Cas1 and Cas2 dimer structures in E. faecalis and E. coli Cas1-Cas2/DNA complexes.
a, Structural analysis of individual Cas1 subunit in the EfaCas1-Cas2/DNA complex. Cas1 consists of a N-term β-sandwich (in yellow circle) and a C-term helical domain (in blue circle). These two domains are connected by a flexible hinge loop (in red circle). b, Superposition of the catalytic (Cas1B, in Cyan) and non-catalytic (Cas1A, in violet) Cas1 subunit in the complex. Note the ~33° hinge motion between NTD and CTD, taking place at the circled region. c, Structural analysis of individual Cas2 subunit in the EfaCas1-Cas2/DNA complex. Monomeric EfaCas2 structure contains a ferredoxin domain. d, Comparison of E. faecalis and E. coli Cas1 in the corresponding Cas1-Cas2/DNA complexes along the 2-fold symmetry axis of the Cas1-NTD dimer. The Cas1-CTD dimer tilts at different angles in these two compex structures. e, EfaCas2 dimerizes at a tilted angle whereas EcoCas2 dimerizes in a juxtaposed fashion (follow the angle between major helices in the dimer). EfaCas2 features long positively charged spikes at its dorsal region, which are inserted into the major grooves of dsDNA for prespacer binding. Overall, the structures of individual Cas1 and Cas2 domains are fairly conserved. The altered overall dimension of the Cas1-Cas2 compex was due to the altered domain orientation at each subunit interface.