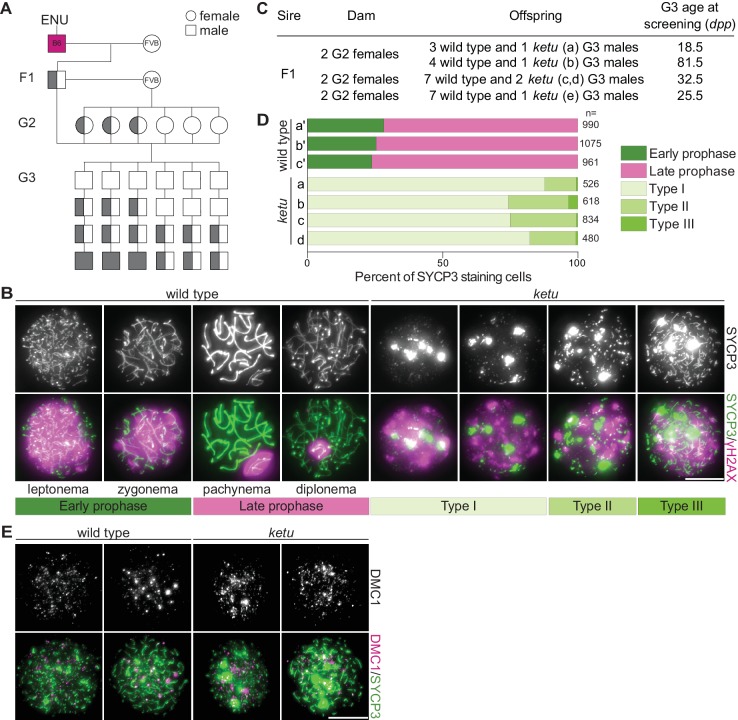
Figure 1. Mice from the ENU-induced mutant line ketu have meiotic defects.
(A) Breeding scheme. Mutagenized males (B6) were crossed to females of a different strain (FVB) to produce founder (F1) males that were potential mutation carriers. Each F1 male was then crossed to wild-type FVB females. If an F1 male was a mutation carrier, half of his daughters (second generation, G2) should also be carriers, so the G2 daughters were crossed back to their F1 sire to generate third-generation (G3) offspring that were potentially homozygous. For a line carrying a single autosomal recessive mutation of interest, one eighth of G3 males were expected to be homozygous. Un-filled shapes represent animals that are wild-type for a mutation of interest, half-filled shapes are heterozygous carriers, and filled shapes are homozygotes. (B) Representative images of squashed spermatocyte preparations immunostained for SYCP3 and γH2AX. Mutant spermatocytes were classified as Types I, II, or III on the basis of SYCP3 patterns. (C) Screen results for the ketu line. The F1 male was harem-bred to six G2 females, yielding 26 G3 males that displayed either a wild-type or ketu (mice a, b, c, d, e) phenotype. (D) Distribution of SYCP3-staining patterns in four G3 ketu mutants (a, b, c, d) and their phenotypically wild-type littermates (a′, b′, c′). Wild-type spermatocytes were classified as either early prophase-like (leptonema or zygonema) or late prophase-like (pachynema or diplonema). Spermatocytes from mutant mice were categorized as described in panel B. The number of SYCP3-positive spermatocytes counted from each animal (n) is indicated and raw data are provided in Figure 1—source data 1. (E) Representative images of squashed spermatocyte preparations immunostained for DMC1 and SYCP3. Scale bars in panels B and E represent 10 μm.