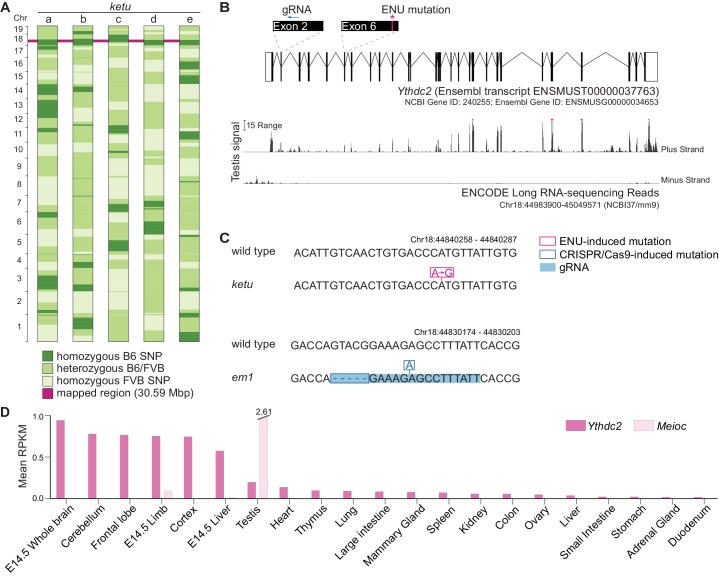
Figure 2. ketu mice harbor a point mutation in Ythdc2.
(A) SNP genotypes of five G3 ketu mutants (a, b, c, d, e; from Figure 1C) obtained using the Illumina Medium Density Linkage Panel. The single 30.59-Mbp region of B6 SNP homozygosity that is shared between mutants is highlighted in pink. (B) Top: Schematic of Ythdc2 (as predicted by Ensembl release 89) showing the locations of the ENU-induced lesion and the gRNA used for CRISPR/Cas9-targeting. Bottom: The density of ENCODE long RNA-sequencing reads (release 3) from adult testis within a window spanning from 3500 bp upstream to 200 bp downstream of Ythdc2. The vertical viewing range is 0–50; read densities exceeding this range are overlined in pink. (C) The ketu and CRISPR/Cas9-induced (em1) alleles of Ythdc2. (D) Ythdc2 and Meioc expression level estimate (mean reads per kilobase per million mapped reads (RPKM) values provided by ENCODE; included in Figure 2—source data 1) in adult and embryonic tissues.