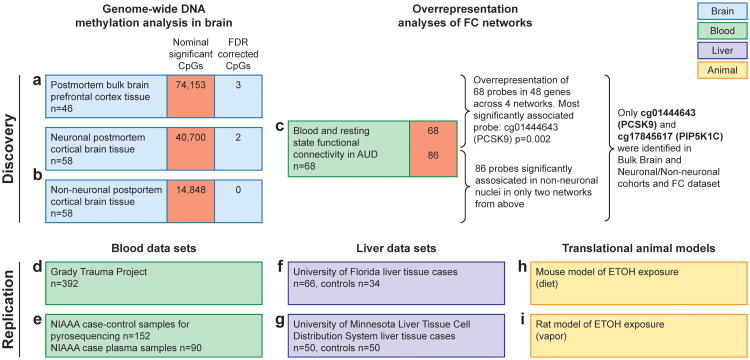
Figure 1. Methylomic profiling approach in AUD using three discovery and six replication data sets identified PCSK9 as main epigenetic target.
A schematic representation of cohorts investigated in this study broken into the discovery phase experiments (a,b,c) that identified PCSK9 association with alcohol use and the replication stage experiments (d-i) including biological target validation in animal models (h,i). Replication experiments were performed in multiple cohorts between blood and liver and data derived from publicly available datasets and direct investigation. For human blood, DNA from individuals who participated in the Grady Trauma Project (GTP) (n= 392) (d) was analyzed for CpG cg01444643 (hg38, chr1: 55039175) (PCSK9CpG1) PCSK9CpG1 was significantly associated with both the binary SCID37 lifetime alcohol abuse (β= -0.007 ± 0.004, F= 3.891, df= 2/325, P= 0.049) and the continuous KMSK Lifetime Alcohol scale38 (β= -0.001 ± 0, F= 4.944, df= 2/306, P= 0.027). In a second human blood cohort (e), we assessed PCSK9 DNA methylation levels with pyrosequencing at the PCSK9CpG1 and an adjacent CpG located at chr1: 55039185 (PCSK9CpG2) in human subjects (n= 90) aged 21-65 with a diagnosis of alcohol dependence and healthy volunteers (n= 62). We observed significantly lower DNA methylation at both CpGs (Student's t-test, PCSK9CpG1: Alcohol Abuse= 76.68 ± 0.05, No Alcohol Abuse= 78.6± 0.064, P= 0.0075; PCSK9CpG2 : Alcohol Abuse= 88.49 ± 0.025, No Alcohol Abuse= 89.28 ± 0.032, P= 0.028). In a liver cohort, we assessed PCSK9 DNA methylation levels at PCSK9CpG1 in DNA derived from individuals with normal livers (n= 34) or primary liver disease tissue arising in the setting of chronic hepatitis B (HBV) or C (HCV) viral infection, alcoholism (ETOH), and other causes (n= 66) (GSE60753) (f). PCSK9 DNA methylation was significantly elevated with alcohol-induced cirrhosis (Alcohol Cirrhosis= 0.54 ± 0.0023, Healthy= 0.49 ± 0.0015, P= 0.0021). Importantly, significant elevations were also observed with cirrhosis induced by hepatitis B (Hep B Cirrhosis= 0.55 ± 0.0079, Healthy= 0.49 ± 0.0015, P= 0.023) and C (Hep C Cirrhosis= 0.57 ± 0.0011, Healthy= 0.49 ± 0.0015, P= 1.2×10-9). Finally, PCSK9CpG1 DNA methylation levels assessed with pyrosequencing in liver tissue samples from liver transplant candidates with alcoholic cirrhosis (n= 50) and healthy controls (n= 47) (g). A significantly higher PCSK9 DNA methylation level was observed in alcoholic cirrhosis cases relative to controls (Student's t-test; Alcohol Abuse= 46.19 ± 1.07, No Alcohol abuse= 37.63 ± 0.89, P= 6.5×10-9). In translational models, mouse liver PCSK9 expression was significantly lower in the alcohol exposure group (h) (Student's t-test; Alcohol Exposure= 0.1 ± 0.011, No Alcohol Exposure= 1.02 ± 0.057, P= 0.0029). A translational rat model (i) was used to assess long-term effects of alcohol on PCSK9 expression in liver.