Abstract
As the HIV/AIDS pandemic still progresses, understanding the mechanisms governing viral transmission as well as protection from HIV acquisition is fundamental. In this context, cohorts of HIV serodiscordant heterosexual couples (SDC) represent a unique tool. The present study was aimed to evaluate specific parameters of innate, cellular and humoral immune responses in SDC. Specifically, plasma levels of cytokines and chemokines, HIV-specific T-cell responses, gp120-specific IgG and IgA antibodies, and HIV-specific antibody-dependent cellular cytotoxicity (ADCC) activity were assessed in nine HIV-exposed seronegative individuals (ESN) and their corresponding HIV seropositive partners (HIV+-P), in eighteen chronically infected HIV subjects (C), nine chronically infected subjects known to be HIV transmitters (CT) and ten healthy HIV− donors (HD). Very low magnitude HIV-specific cellular responses were found in two out of six ESN. Interestingly, HIV+-P had the highest ADCC magnitude, the lowest IgA levels and the highest IgG/IgA ratio, all compared to CT. Positive correlations between CD4+ T-cell counts and both IgG/IgA ratios and %ADCC killing uniquely distinguished HIV+-P. Additionally, evidence of IgA interference with ADCC responses from HIV+-P and CT is provided. These data suggest for the first time a potential role of ADCC and/or gp120-specific IgG/IgA balance in modulating heterosexual transmission. In sum, this study provides key information to understand the host factors that influence viral transmission, which should be considered in both the development of prophylactic vaccines and novel immunotherapies for HIV-1 infection.
Keywords: HIV-1, Serodiscordant couples, ADCC, HIV-1 transmission
Highlights
-
•
The evaluation of different immune parameters in HIV serodiscordant couples helped identify factors shaping transmission.
-
•
Innate and cellular immune responses were apparently not involved in this scenario.
-
•
HIV-specific ADCC, IgA titer and IgG/IgA balance were identified as factors involved in modulating viral transmission.
The existence of individuals that remain HIV negative despite being repeatedly exposed to the virus has long been described. To date, only homozygosis for a 32-base pair deletion in the ccr5 gene has been consistently shown to be a determinant of HIV resistance. Still, subjects bearing the WT ccr5 gene have also been described as resistant or less susceptible to HIV. Thus, other mechanisms must be involved in this phenomenon. The results presented here postulate ADCC and IgG/IgA ratio as potential mechanisms involved in modulating HIV transmission in the context of serodiscordant couples and inspire further investigations.
1. Introduction
The pandemic of HIV/AIDS is still a major public health concern worldwide (Frank and Witten, 2016). Highly active antiretroviral therapy (HAART) can diminish viral loads (VL) to undetectable levels, reducing not only mortality but also transmission rates, with the subsequent impact on a population level (Hull et al., 2014). However, it does not cure the infection. Moreover, an effective vaccine has not been developed yet (Deeks et al., 2016).
It is well known the fact that not all HIV-infected individuals progress to AIDS at the same rate. For instance, there is a peculiar population of HIV+ subjects able to maintain undetectable VL for > 10 years in the absence of HAART and AIDS-related diseases. These subjects are known as Elite Controllers (ECs). Even though the correlates of protection in these individuals have not been fully identified, it has been suggested that genetic (presence of HLA-B*57, HLA-B*27, HLA-B*13 and HLA-B*58.01) and immune (CD8+ T-cell response) host factors could be involved (McDermott and Koup, 2012, Gonzalo-Gil et al., 2017). Similarly, recent evidence indicated that humoral immunity could mediate protection in this group (Lambotte et al., 2009, Ackerman et al., 2016).
In the same line, there are other individuals that, despite having being exposed to the virus for long periods of time, are resistant to HIV infection (Rowland-Jones and McMichael, 1995). This phenomenon was observed among (i) heterosexual partners of HIV infected people (Ranki et al., 1989, Langlade-Demoyen et al., 1994), (ii) sex workers (Rowland-Jones et al., 1995, Fowke et al., 1996), (iii) men who have sex with men, (iv) intravenous drug users, (v) exposed uninfected infants (Rowland-Jones et al., 1993, Cheynier et al., 1992) and, (vi) health-care workers (Clerici et al., 1994, Pinto et al., 1995). The mechanisms that allow these individuals to be “protected” from the virus are still unknown, although several hypotheses have been proposed. To date, only homozygosis for a 32-base pair deletion in the gene encoding the CCR5 protein (CCR5Δ32), the major coreceptor used by viral isolates most frequently associated with transmission events (R5-tropic HIV-1 variants) has been consistently shown to be a determinant of HIV resistance (Liu et al., 1996). Still, subjects bearing the WT ccr5 gene have been described as resistant or less susceptible to HIV infection. Thus, unraveling this mystery and understanding the underlying mechanism could help in the development of novel therapies and even a vaccine.
Sexual transmission is currently the major route of HIV infection worldwide accounting for > 80% of new infections (Hladik and McElrath, 2008). By definition, a HIV serodiscordant couple (SDC) is a couple in which one partner is HIV-positive and the other is HIV-negative. SDC cohorts may be the most relevant groups for identifying correlates of protection affecting sexual transmission. The first evidence of resistance to infection in spite of exposure in this kind of cohorts appeared in 1989, when T-cell responses to HIV proteins were observed in the seronegative partners, later defined as exposed seronegative (ESN) individuals (Ranki et al., 1989). Since then, cellular, humoral and innate immune responses in ESN subjects have been studied (review in (Piacentini et al., 2008)). Remarkably, there are certain aspects of the immune response that have been recently associated with protection from disease progression but have not been investigated in the scenario imposed by SDC yet. This is the case of antibody-dependent cellular cytotoxicity (ADCC). HIV-specific ADCC-mediating antibodies have been found in plasma of HIV-infected individuals (Forthal et al., 2001, Dugast et al., 2014, Cereb et al., 1995), in cervicovaginal fluids (Battle-Miller et al., 2002), breast milk (Mabuka et al., 2012) and semen (Parsons et al., 2016) of infected subjects. Several reports suggest that ADCC-mediating antibodies might protect infected individuals from disease progression (Lambotte et al., 2009, Thobakgale et al., 2012, Baum et al., 1996, Chung et al., 2011). Recently, our group demonstrated that gp120-specific IgA is a plasma factor capable of modifying the magnitude of IgG-mediated ADCC in HIV infection, possibly abrogating its protective role (Ruiz et al., 2016). The presence of antibodies capable of mediating ADCC at sites of viral entry raises the possibility that these antibodies could modulate viral transmission, probably by inhibiting or decreasing transmission rates. In this line, it was reported that passively acquired ADCC activity in infants born to HIV-infected mothers was not associated with protection but with reduced mortality (Milligan et al., 2015).
In sum, HIV transmission is a complex phenomenon (Dale et al., 2013). The group of SDC is a valuable set of subjects that may help us to study not only correlates of protection in ESN, but also the existence of possible factors involved in viral transmission. Here, we aimed to evaluate different aspects of innate, cellular and humoral immune responses in a cohort of SDC. For this, plasma levels of 39 cytokines and chemokines, HIV-specific T-cell responses, gp120-specific IgG and IgA antibodies, and HIV-specific ADCC activity were evaluated in ESN from SDC and their respective HIV+ partners (HIV+-P). Also, the same parameters were evaluated in chronically infected HIV+ subjects (C), chronically infected subjects known to be HIV transmitters (CT) and healthy donors (HD), for comparison purposes. We hypothesized that the joint evaluation of the aforementioned parameters will shed light on possible immune correlates of protection in ESN as well as possible factors affecting viral transmission. Interestingly, ADCC magnitude, gp120-specific IgA titers and gp120-specific IgG/IgA ratio appeared as factors able to discriminate HIV+-P, CT and C subjects. This warrants further studies on the role of gp120-spcific IgA, ADCC-mediating antibodies and the relation of these two factors in modulating viral transmission.
2. Materials and Methods
2.1. Study Groups
The following study groups were enrolled (Table 1): Nine serodiscordant heterosexual couples (SDC), eighteen chronically infected subjects (C) and nine chronically infected subjects known to be HIV transmitters (CT). Also, ten healthy HIV-seronegative donors (HD) were enrolled as a control group. SDC were enrolled based on self-reporting being a couple for a period longer than three years and having unprotected sex during that period. The HIV+ individual of the couple (hereinafter HIV+ positive partner (HIV+-P)) self-reported being off-HAART throughout that period and had detectable (> 50 HIV RNA copies/ml plasma) viral load (VL) at the moment of enrollment. It is important to highlight that these subjects were not followed up by our team, and SDC were enrolled prior to the development of the HIV treatment guidelines which indicate that all individuals with a negative partner should receive treatment as prevention. Seronegative status of the HIV negative subject (from now on exposed seronegative partner, ESN) was corroborated at the moment of enrollment.
Table 1.
Data corresponding to enrolled subjects per study group.
| Subject ID | Sex | Age (years) | Time of exposurea (years) | Plasma viral load (VL)b |
CD4 + T cell countc (cells/μl) | HLA |
Viral subtyped | ||
|---|---|---|---|---|---|---|---|---|---|
| (HIV RNA copies/ml) | Log10 (copies/ml) | A | B | ||||||
| Group SDC: serodiscordant couples (N = 9)e | |||||||||
| HIV+-P1 | F | 45 | 6 | 36,364 | 4.56 | 530 | 02/25 | 8/51 | BF |
| ESN1 | M | 40 | < 50 | < 1.7 | 1208 | 02/29 | 44CXHZ/7 | − | |
| HIV+-P2 | M | 47 | 10 | 95,821 | 4.98 | 41 | 02/08 | 48/35 | BF |
| ESN2 | F | 43 | < 50 | < 1.7 | 634 | 01/32 | 44CXJC/18 | − | |
| HIV+-P3 | M | 37 | 3 | 21,758 | 4.33 | 370 | NA | NA | C |
| ESN3 | F | 32 | < 50 | < 1.7 | 904 | 02/34 | 40CXJJ (61)/14 | − | |
| HIV+-P4 | F | 33 | 5 | 177 | 2.24 | 386 | 11/68 | 35/08 | NA |
| ESN4 | M | 26 | < 50 | < 1.7 | 850 | 02/24 | 15CXHX (62)/07 | − | |
| HIV+-P5 | M | 34 | 5–7 | 901 | 2.9 | 160 | 02/02 | 39/39 | NA |
| ESN5 | F | 73 | < 50 | < 1.7 | 672 | 01/02 | 8/44CXJC | − | |
| HIV+-P6 | M | 30 | 10 | 364,928 | 5.56 | 21 | NA | NA | BF |
| ESN6 | F | 31 | < 50 | < 1.7 | 713 | NA | NA | − | |
| HIV+-P7 | M | 41 | 5 | 15,867 | 4.2 | 333 | 68/36 | 07/44CXJC | BF |
| ESN7 | F | 43 | < 50 | < 1.7 | 606 | 02/11 | 07/38 | − | |
| HIV+-P8 | M | 40 | > 3 | 81 | 1.9 | 259 | 2/24 | 25/29 | NA |
| ESN8 | F | 25 | < 50 | < 1.7 | 1214 | 40/35 | 50/36 | − | |
| HIV+-P9 | F | 36 | > 3 | 465 | 3.05 | 256 | 01/33 | 45/52 | NA |
| ESN9 | M | 26 | < 50 | < 1.7 | 497 | 32/32 | 8/38 | − | |
| Group C: chronics (N = 18) | |||||||||
| C01 | F | 21 | − | 85,947 | 4.9 | 25 | 11/24 | 35/51or78f | BF |
| C02 | M | 22 | − | 18,580 | 4.2 | 511 | 1/3 | 7/37 | B |
| C03 | M | 49 | − | 43,436 | 4.6 | 479 | 31/31 | 15/39 | |
| C04 | M | 28 | − | 555 | 2.7 | 555 | 2/3 | 44/51 | BF |
| C05 | M | 36 | − | 1288 | 3.1 | 431 | NA/NA | NA/NA | C |
| C06 | F | 27 | − | 1817 | 3.2 | 689 | 11/11 | 35/40 | |
| C07 | M | 47 | − | 11,026 | 4.0 | 585 | 2/2 | 44/28 | |
| C08 | F | 38 | − | 22,475 | 4.3 | 143 | 2/2or25f | 35/55 | B |
| C09 | F | 29 | − | 14,784 | 4.3 | 139 | 26/26 | 44/57 | BF |
| C10 | F | 24 | − | 253,167 | 5.4 | 5 | 31/31 | 39/47 | BF |
| C11 | M | 23 | − | 45,199 | 4.7 | 282 | 11/11 | 18/35 | |
| C12 | F | 50 | − | 3093 | 3.5 | 227 | 2/33 | 15/39 | |
| C13 | F | 24 | − | 1286 | 3.1 | 1071 | 2/3 | 35/44 | BF |
| C14 | M | 26 | − | 22,060 | 4.3 | 293 | 2/2 | 7/35 | BF |
| C15 | M | 33 | − | 4698 | 3.7 | 408 | 31/31 | 8/48 | BF |
| C16 | M | 40 | − | 25,402 | 4,4 | 692 | 2/36 | 7/42 | B |
| C17 | M | 73 | − | 58,410 | 4,8 | 575 | 2/2 | 39/52 | B |
| C18 | M | 47 | − | 710 | 2,9 | 678 | 31/68 | 44/51 | BF |
| Group CT: chronics with documented transmission event (N = 9) | |||||||||
| CT01 | M | 39 | − | 25,599 | 4.4 | 223 | 31/X | 1508 (75)/35 | B |
| CT02 | M | 29 | − | 50,563 | 4.7 | 269 | 32/26 | 38BHS/44CXHZ | F |
| CT03 | M | 38 | − | 383,841 | 5.5 | 8 | 2/68 | 7CXPJ/15CXHW | B |
| CT04 | F | 40 | − | 25,899 | 4.4 | 352 | 2/24 | 15YCK/1517 | BF |
| CT05 | F | 29 | − | 1904 | 3.2 | 357 | NA/NA | NA/NA | BFC |
| CT06 | M | 30 | − | 264,322 | 5.4 | 227 | 11/3001 | 18/35 | BF |
| CT07 | M | 32 | − | 30,020 | 4.4 | 238 | NA/NA | NA/NA | BF |
| CT08 | M | 28 | − | 7666 | 3.8 | 294 | NA/NA | NA/NA | B |
| CT09 | M | 33 | − | 19,250 | 4.2 | 495 | NA/NA | NA/NA | BF |
F: Female, M: Male. NA; Not available.
Estimated time based on self-reported data on clinical questionnaire.
Versant HIV-1 RNA 3.0 assay, Siemens. Lower and upper detection limits was 50 and 500,000 RNA copies/ml, respectively (1.7 and 5.7 log10).
Flow cytometry double platform, FACSCanto, BD Biosciences.
Viral subtype determined by pol gene sequencing.
Both individuals belonging to one couple are shown in side by side lines and are indicated with the same number. HIV+-P: HIV Positive Partner, ESN: Exposed seronegative subject.
Could not be exactly determined.
C and CT were defined as subjects infected for > 3 years, with detectable VL and HAART naïve. In addition, CT had at least one transmission event documented by self-reported, clinical and phylogenetic data (Damilano G, unpublished). Samples from HD were obtained from > 18 year-old voluntary blood donors who completed and passed a survey on blood donation which specifically excludes persons who had been exposed to HIV; and were screened for serological markers before being accepted as donors.
Blood samples from HIV-infected individuals, uninfected sexual partners from HIV-infected individuals, and healthy donors were obtained for this study. Prior to enrollment, the study was reviewed and approved by the Comité de Ética Humana, Facultad de Medicina, Universidad de Buenos Aires. All participants provided written informed consent accepting to participate in this study.
2.2. Samples
Plasma and peripheral blood mononuclear cells (PBMCs) were obtained from ESN and HIV+-P (both individuals from the SDC), HD and C. For CT only plasma samples were obtained with the purpose of being used as control group for ADCC assays. In all cases, blood samples were collected and centrifuged to separate plasma, which was stored at − 80 °C until use. For ADCC assays, plasma samples were first diluted (10-fold in RPMI medium) passed through a 0.2 μm-pore filter and heat-inactivated (1 h, 56 °C). PBMCs were isolated by Ficoll-Hypaque density gradient centrifugation (GE Healthcare, UK) and cryopreserved for subsequent assays. PBMCs from one HIV negative donor were isolated, cryopreserved and used as effector cells in ADCC assay. Cells from the same donor were used in all assays to avoid bias from donor to donor. Plasma VL (branched-DNA, Versant HIV-1 RNA 3.0 assay; Siemens Healthcare, UK) and CD4+ T-cell count (flow cytometry double platform, BD FACSCanto; BD Biosciences, USA) were assessed in all subjects.
2.3. HIV Subtype Determination
HIV-1 RNA was extracted from 200 μl of stored plasma using the viral RNA extraction Kit (Purelink viral RNA/DNA Mini kit, Invitrogen-Life Technologies). Afterwards, pol gene was amplified by RT-PCR as described before (Dilernia et al., 2013). Amplicons were purified and sequenced using the Big Dye Terminator sequencing kit v3.1 (Applied Biosystems, USA) on an automated sequencer (Applied Biosystems DNA sequencer 3500). Nucleotide sequences were analyzed and manually adjusted using Sequencher v5.1 software (Gene Codes Co.). The partial pol sequences were aligned independently of each other with HIV-1 references sequences available in Los Alamos Database (http://www.hiv.lanl.gov/content/sequence/HIV/mainpage.html). Alignments were performed with MAFFT software included in http://www.hiv.lanl.gov/content/sequence/VIRALIGN/viralign.html (Katoh and Standley, 2014). Subtype determination was performed separately for each sequence by following different methods: jumping profile Hidden Markov Model (jpHMM) (Schultz et al., 2009), recombination detection program (RDP4) (Martin et al., 2015) and recombination analysis using cost optimization (RECCO) (Maydt and Lengauer, 2006). Subtype was assigned when at least 2 out of 3 methods used were coincident.
2.4. HLA Typing and CCR5 Characterization
Total DNA was extracted from 2 × 106 PBMCs using QIAamp DNA kit (Qiagen GmbH, Germany). HLA-A and HLA-B loci were typed, at the two-digit level, using PCR-single-stranded oligonucleotide probes as described previously (Cereb et al., 1995, Fernandez-Vina et al., 1997). CCR5-Δ32 deletion was identified by differences in PCR product sizes as in (Coloccini et al., 2014). A band of 184 bp was representative of the normal allele while a band of 152 pb corresponded to CCR5Δ32 allele.
2.5. Quantification of Soluble Plasma Factors
Simultaneous determination of the following 39 cytokines and chemokines was performed using Luminex technology (MILLIPLEX MAP Human Cytokine/Chemokine, Merck Millipore, Billerica/Massachusetts, USA): EGF, Eotaxin, FGF-2, Flt-3 Ligand, Fractalkine, G-CSF, GM-CSF, GRO, IFN-α2, IFN-γ, IL-1α, IL-1β, IL-1rα, IL-2, IL-3, IL-4, IL-5, IL-6, IL-7, IL-8, IL-9, IL-10, IL-12 (p40), IL-12 (p70), IL-13, IL-15, IL-17, IP-10, MCP-1, MCP-3, MDC (CCL22), MIP-1α, MIP-1β, sCD40L, sIL-2Rα, TGF-α, TNF-α, TNF-β, VEGF. Samples were processed and analyzed as described by Giavedoni et al. (Giavedoni, 2005).
2.6. HIV-specific Cellular Immune Responses
The magnitude and specificity of the HIV-specific cellular immune response was screened by IFN-γ ELISPOT as described previously (Turk et al., 2008, Turk et al., 2013). Briefly, cryopreserved PBMCs were thawed in complete RPMI medium (RPMIc; RPMI 1640 [Gibco BRL], 10% fetal bovine serum [FBS; Gibco BRL], 2 mM l-glutamine [Gibco BRL], 100 U/ml penicillin [Gibco BRL], 100 μg/ml streptomycin [Gibco BRL], 10 mM HEPES [Gibco BRL]) supplemented with 50 U/ml DNase I (Benzonase nuclease; Sigma-Aldrich) and then rested overnight in DNase-free medium. Rested PBMCs with > 95% viability were plated on sterile 96-well plates (MultiScreen IP plates; Millipore), previously coated with mouse anti-human IFN-γ monoclonal antibody (BD Biosciences) at 105 cells/well, and peptide pools (spanning HIV proteins Nef, Gag (three pools) and Env (five pools)) were added at a final concentration of 2 μg/ml of each peptide (Potential T-cell epitope (PTE) peptide panels corresponding to Nef, Gag, and Env proteins were obtained from the NIH AIDS Reagent Program (Malhotra et al., 2007)). The PTE peptides are 15 amino acids (aa) in length and contain naturally-occurring 9-aa sequences that are potential T-cell determinants embedded in the sequences of multiclade circulating HIV-1 strains worldwide. Final DMSO concentration was always lower than 0.7%. Negative (peptide-free medium plus 0.5% DMSO), Cytomegalovirus, Epstein Bar and Influenza (CEF) peptide pool (2 μg/ml) of each peptide, NIH AIDS Reagent Program (Currier et al., 2002), and phorbol myristate acetate (PMA)-ionomycin (5 ng/ml PMA plus 500 ng/ml ionomycin; Sigma-Aldrich) controls were assayed for each subject. Alternatively, 105 and 106 cells/well were tested for ESN samples. Plates were developed using biotinylated anti-human IFN-γ monoclonal antibody, streptavidin-peroxidase complex, and the 3-amino-9-ethylcarbazole (AEC) substrate reagent set (BD Biosciences). Plates were scanned on an ImmunoSpot reader (Cellular Technology Ltd.). Specific spot count was calculated using the ImmunoSpot software. Results were expressed as spot-forming units (SFU)/106 PBMCs after subtracting the negative-control values. Gag and Env responses represented the sum of the responses obtained for each of its corresponding 3 and 5 pools, respectively. Thresholds for positive responses were defined as at least 50 SFU/106 PBMCs or as mean SFU greater than three times the mean SFU of the negative-control wells, whichever was higher.
When stated, a modified protocol denoted as ¨expanded ELISPOT¨ was used to increase sensitivity. For this, thawed and ON rested PBMCs were cultured in 12-well plates at a density of 2 × 106 cells/ml in complete RPMI medium supplemented with 1800 U/ml IL-2 and in the presence of 2 μg/ml of the corresponding HIV peptide pool, for 14 days (Goonetilleke et al., 2006, Rodriguez et al., 2009). Medium was replaced every 72 h with freshly prepared IL-2. At day 14, ELISPOT was performed as described above.
2.7. ELISA for Env-specific Plasma IgG and IgA
Gp120-specific IgG concentration and gp120-specific IgA titers were determined by ELISA as described before (Ruiz et al., 2016). 96-well flat-bottomed half-area plates (GreinerBio-One, Germany) were coated with 25 ng/well of gp120BAL. For gp120-specific IgG quantitation, 25 μl/well of 1/10000 (C, CT and HIV+-P) or 1/10 (ESN and HD) plasma dilutions were dispensed in triplicate. A standard curve was constructed, consisting in two-fold serial dilutions of the anti-gp120 2G12 monoclonal antibody starting at 24 ng/ml. IgG detection was performed using an anti-human IgG antibody labeled with horseradish peroxidase (HRP, Sigma-Aldrich, USA) and developed with TMB-ELISA solution (BD Biosciences, USA). Absorption at 450 nm was read on Multiskan EX Microplate Reader (Thermo/Labsystems). IgG concentration was extrapolated from the standard curve and multiplied by the dilution factor. Gp120-specific IgA levels were determined by end-point titration. Two-fold serial dilutions of plasma were prepared, starting at 1:20. Secondary antibody was an anti-human IgA-HRP (Sigma-Aldrich, USA). Plates were developed and read as described for IgG. End-point IgA titer was defined as the reciprocal of the highest plasma dilution at which the average OD value was ≥ 2-fold the average OD value of control wells. Sera from HIV-negative subjects were tested as controls.
2.8. Rapid Fluorometric ADCC (RFADCC) Assay
The assay was performed as described by others (Gomez-Roman et al., 2006) and later adapted by our group (Ruiz et al., 2016). Briefly, CEM-NKR target cells (AIDS Research and Reference Reagent Program (Howell et al., 1985)) were double-stained with PKH-26 (Sigma-Aldrich, USA) and CFSE (Carboxyfluoresceindiacetatesuccinimidyl ester; Molecular Probes, USA) and coated with recombinant gp120BAL (AIDS Research and Reference Reagent Program, HIV-1BaLgp120 from DAIDS, NIAID). After one hour, cells were dispensed in U-bottom 96-well plates (5000 cells/well) together with different dilutions of inactivated plasma, in triplicate. After 15 min at room temperature, effector cells (thawed and overnight rested PBMCs) were added at an effector/target ratio of 50:1. Plates were centrifuged and incubated for 4 h at 37 °C. Cells were washed, fixed, acquired in a FACSCanto flow cytometer (BD Biosciences, USA) and analyzed using the FACSDiva v6.1.3 software (BD Biosciences) or FlowJO v10 (Data Analysis Software, LLC). Target cells were initially gated in a forward scatter (FSC)-versus side scatter (SSC) plot and subsequently gated on a SSC versus PKH-26 plot. Then, a PKH-26 versus CFSE plot was generated to determine target-cell killing (%ADCC killing) which was calculated as the proportion of cells that remained PKH-26high but had lost the viability dye (CFSEneg). Results are presented as the media of triplicate conditions and background-subtracted (%ADCC killing for uncoated target cells). Threshold for positive responses for a given plasma dilution was defined as the mean of the background plus three SD.
Coating and saturation of target cells were verified by flow cytometry with anti-gp120 monoclonal antibody 2G12 (AIDS Research and Reference Reagent Program) followed by staining with an anti-human IgG-APC.
2.9. Depletion of Plasma IgA
Bulk IgA was removed from plasma obtained from one representative HIV+-P and one representative CT individual using the Pierce Immunoprecipitation kit (Thermo Scientific, USA) as described previously (Ruiz et al., 2016). Briefly, diluted and filtered plasma samples were added to anti-IgA coupled columns, incubated overnight at 4 °C and eluted by centrifugation. Flow-through was collected and stored for further analysis of ADCC responses. Plasma passed through columns coupled with an isotype-matched control antibody was used as control (non-depleted plasma). Specific depletion of IgA was confirmed by dot-blot and ELISA.
2.10. Data Analysis
Statistical analyses were performed using GraphPad Prism 6 Software. Data was expressed as median values with interquartile ranges (IQ25-75) and analyzed by non-parametric methods unless otherwise stated. Outliers were identified by the Grubbs' method using a two-tailed α = 0.05. Mann-Whitney test or Kruskal-Wallis plus Dunn's post-test were used to compare variables between two or multiple groups, respectively. Correlations were determined using Spearman's rank test. All tests were considered significant when p value was < 0.05. When analyzing differences in soluble plasma molecules between groups, p values were adjusted for multiple comparisons using a false discovery rate (FDR) procedure, according to the Benjamini & Hochberg method, with R Project software version 3.3.3. Adjusted p values were considered significant when < 0.1.
3. Results
3.1. Study Subject Description
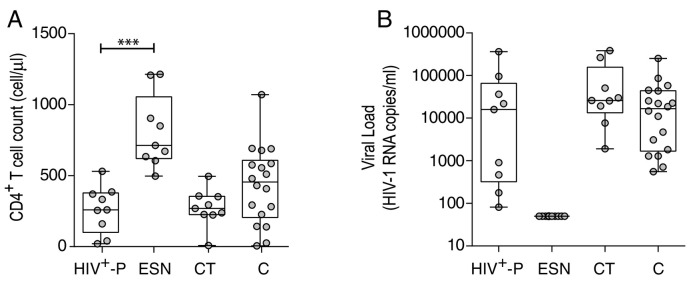
Nine exposed HIV seronegative individuals (ESN) and their respective HIV+ partners (HIV+- P) were enrolled. All SDC were heterosexual. Among them, three HIV+- P were female and six were male. Additionally, eighteen chronically infected subjects (C), nine chronically infected subjects known to be HIV transmitters (CT) and ten healthy HIV-seronegative donors (HD) were enrolled for this study. Details for each subject are described in Table 1. Plasma viral load (VL) and CD4+ T-cell count were determined in all groups, except for HD. HIV+-P median CD4+ T-cell count was 259 cell/μl and median VL was 15,867 HIV RNA copies/ml (Fig. 1). As expected, all ESN had undetectable VL (according to their seronegative status at the moment of enrollment, not shown). The ESN median CD4+ T-cell count was 713 cells/μl, which was significantly higher compared to the HIV+-P group (p < 0.0001, Fig. 1). C and CT had a median of 455 and 269 CD4+ T-cells/μl respectively, and their median VL were 16,682 and 25,899 respectively (Fig. 1). No significant differences were found among the C, CT and HIV+-P groups regarding VL and CD4+ T-cell counts. It is worth noting that some HIV+-P could be considered as “viremic controllers” due to their relative low VL (< 2000 RNA copies/ml). Reduced VL has been directly associated to transmission risk thus these subjects could be less prone to transmit the virus. However, the fact that all HIV+-P had detectable plasma VL and that even high VL subjects are represented within this group indicates that resistance to HIV infection in enrolled ESN might be determined also by other host and/or viral factors.
Fig. 1.
CD4+ T-cell counts (A) and plasma viral loads (VL, B) of enrolled HIV subjects per study group. Inclusion criteria for subjects enrolled are explained in Materials and Methods. Horizontal lines stand for median values. HIV+-P: HIV positive partners, ESN: Exposed HIV seronegative individuals, CT: Chronically HIV transmitters subjects C: Chronics. P values were calculated using a Mann-Whitney U test. Asterisks denote different p values: ***, p < 0.005,
Viral subtypes were determined in most of the HIV+ individuals enrolled for this work and shown in Table 1. The frequency of each of these viral variants within our study group was in line with the frequency observed at the population level in our region (Dilernia et al., 2013). In the case of ESN, the presence of protective HLA alleles (HLA B*57, HLA-B*27, HLA-B*13 and HLA-B*58.01) was tested and excluded (Table 1). Similarly, no ESN presented the CCR5Δ32 deletion.
3.2. Macrophage Derived Chemokine (MDC) was Modestly Elevated in Plasma From ESN Compared to HD While sCD40L was Lower in HIV+-P Compared to Chronics
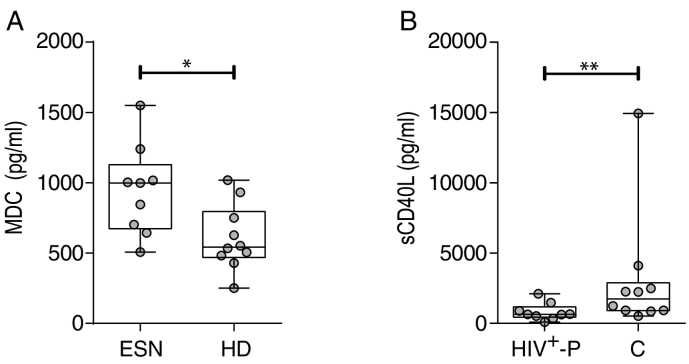
In order to evaluate specific parameters of immune responses in HIV SDC, we first assessed plasma levels of 39 cytokines and chemokines by Luminex technology in both partners of SDCs (ESN and HIV+-P), in a subset of twelve C and in ten HD. In all cases, ESN were compared with HD and HIV+-P with C. Plasma levels of the macrophage-derived chemokine (MDC) were significantly elevated in ESN, compared to HD (median 998 and 543 pg/ml, respectively; p = 0.0220, Fig. 2A), even at levels comparable to those of HIV-infected groups (median 1150 and 849 pg/ml for HIV+-P and C, respectively, p ≥ 0.05). No other significant differences were obtained when comparing ESN with HD. On the other hand, sCD40L, an inflammation biomarker associated with AIDS progression (Sipsas et al., 2002), was detected in significantly lower levels in HIV+-P compared to C (median 647 and 2241 respectively; p = 0.0097, Fig. 2B). Even when removing one outlier identified in the dataset corresponding to C, the difference with the HIV+-P remained statistically significant (p = 0.0172). Moreover, sCD40L levels obtained for HIV+-P were not different from the levels observed in ESN and HD (median 563 and 640 pg/ml, respectively). However, these differences were lost when applying the FDR procedure.
Fig. 2.
Differential plasma cytokine/chemokine levels among groups. A. Macrophage-derived chemokine (MDC) levels (pg/ml) in plasma from exposed HIV seronegative individuals (ESN) versus healthy donors (HD). B. sCD40L levels (pg/ml) in plasma from HIV positive partners (HIV+-P) versus Chronics (C). P values were calculated using a Mann-Whitney U test. Asterisks denote different p values: *, p < 0.05, **, p < 0.001. After applying the FDR procedure, adjusted p value was > 0.1.
Thus, no major difference was observed among soluble cytokines and chemokines when comparing plasma obtained from HD and ESN or HIV+-P and C. Out of the 39 cytokines evaluated, only MDC and sCD40L were significantly different in ESN (compared to HD) and HIV+-P (compared to C) respectively. Even when MDC was described to have HIV suppressive activity (Pal et al., 1997) and sCD40L was reported to be elevated in progressive HIV-1 infection, our data set lacks power to identify these molecules as potential factors involved in modulating HIV transmission events.
3.3. Only a Minor Proportion of ESN Showed HIV-specific Cellular Response and it was of Very Low Magnitude
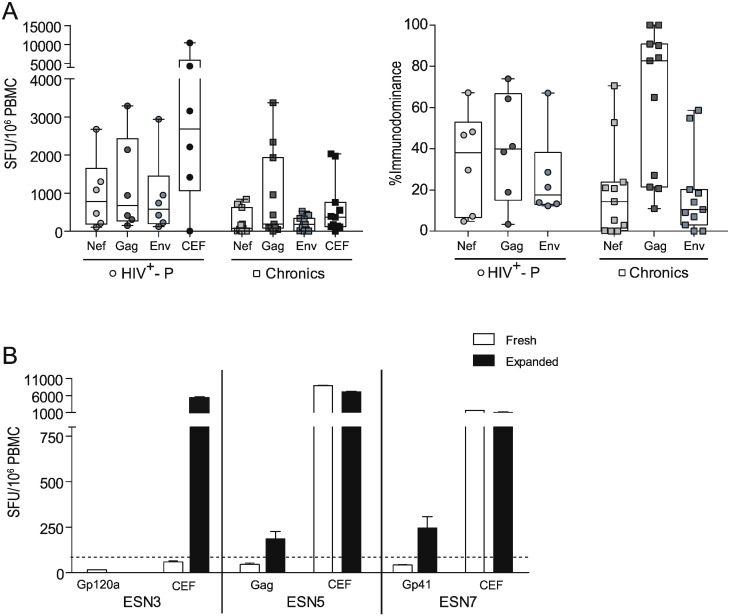
Next, HIV-specific T-cell responses directed towards Nef, Gag and Env proteins were evaluated by ELISPOT in six out of nine SDC. A conventional eighteen-hour stimulation ELISPOT protocol was first used. No significant differences were observed between HIV+-P and C either in magnitude (Fig. 3A, left panel) or immunodominance (Fig. 3A, right panel) of the cellular HIV-specific response. The HIV+-P group showed a slightly higher magnitude for all antigens, even for the CEF control peptide pool. Also, it was observed that the response was broader than in C, which showed a Gag-concentrated response. However, these observations were all trends and no statistically significant differences were found.
Fig. 3.
ELISPOT screening of HIV-specific T-cell response magnitude and breadth in HIV+-P, ESN and Chronics. A. Magnitude (SFU/106 PBMC, left panel) and breadth (contribution of each antigen relative to the total HIV response, right panel) of the HIV-specific T-cell responses in HIV+-P and Chronics. In the case of the magnitude, results corresponding to the CEF control peptide pool (Cytomegalovirus, Epstein Bar, Influenza) are also shown. Horizontal lines within boxes represent the median values. Whiskers extend from min to max values. B. Magnitude (SFU/106 PBMC) of the HIV-specific and CEF-specific responses in three ESN individuals, obtained in the “fresh” (conventional) and “expanded” (14-day stimulation) ELISPOT protocol. Bars represent the media ± SD from triplicate conditions.
In the conventional ELISPOT protocol, three ESN (ESN3, ESN5 and ESN7) showed detectable responses but below the assay cut-off (15, 47 and 40 SFU/106 PBMCs, Fig. 3B). We reasoned that this could be specific responses but of very low magnitude. To address this possibility, a 14-day stimulation protocol (expanded protocol) was applied. Expanded cells from ESN5 and ESN7 showed a significant 4- and 6-fold increase, respectively, in the HIV-specific response (Fig. 3B). Conversely, no response was observed in ESN3 after the expansion with the corresponding HIV peptide pool. However, expansion with the CEF peptide pool in this subject resulted in a 95-fold increase in the magnitude of the CEF-specific response, compared to the fresh protocol, which indicated that the cells were viable and responsive to CEF but not to HIV peptides. Additionally, the 14-day stimulation protocol was tested in a subset of five HDs against all HIV peptide pools and the control CEF pool. No positive HIV-specific responses were found, further confirming the specificity of this protocol (not shown).
Overall, no significant difference was observed in the HIV-specific cellular immune response between HIV+-P and C. Out of the 6 ESN tested, only two had HIV-specific cellular immune responses which were of very low magnitude, confirmed by an expanded ELISPOT protocol. This could be an indication of continuous exposure to the virus.
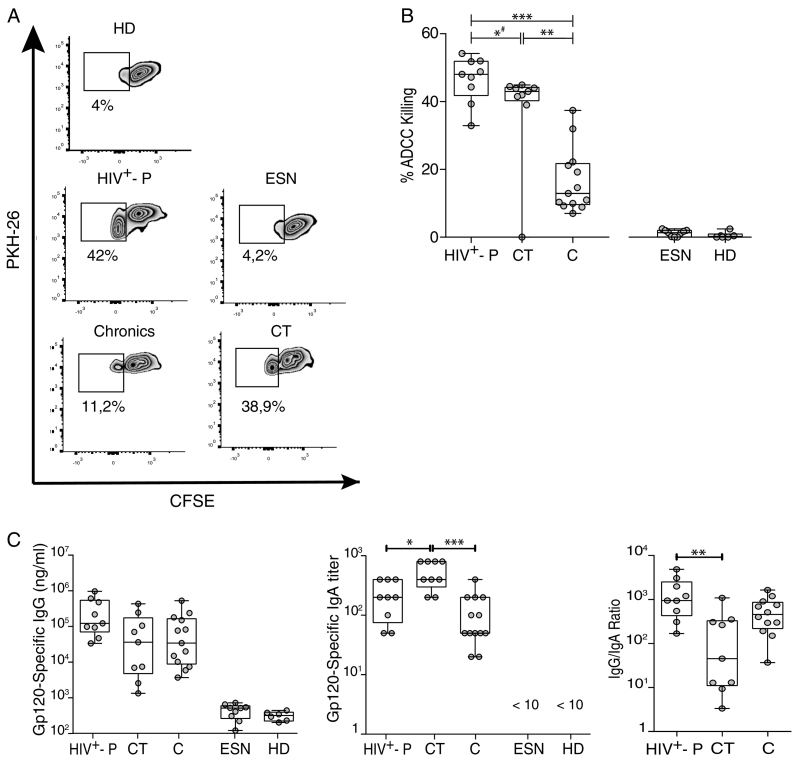
3.4. Most HIV+- P had Higher ADCC Responses Compared to Chronics and CT and Also Statistically Higher gp120-specific IgG/IgA Ratios
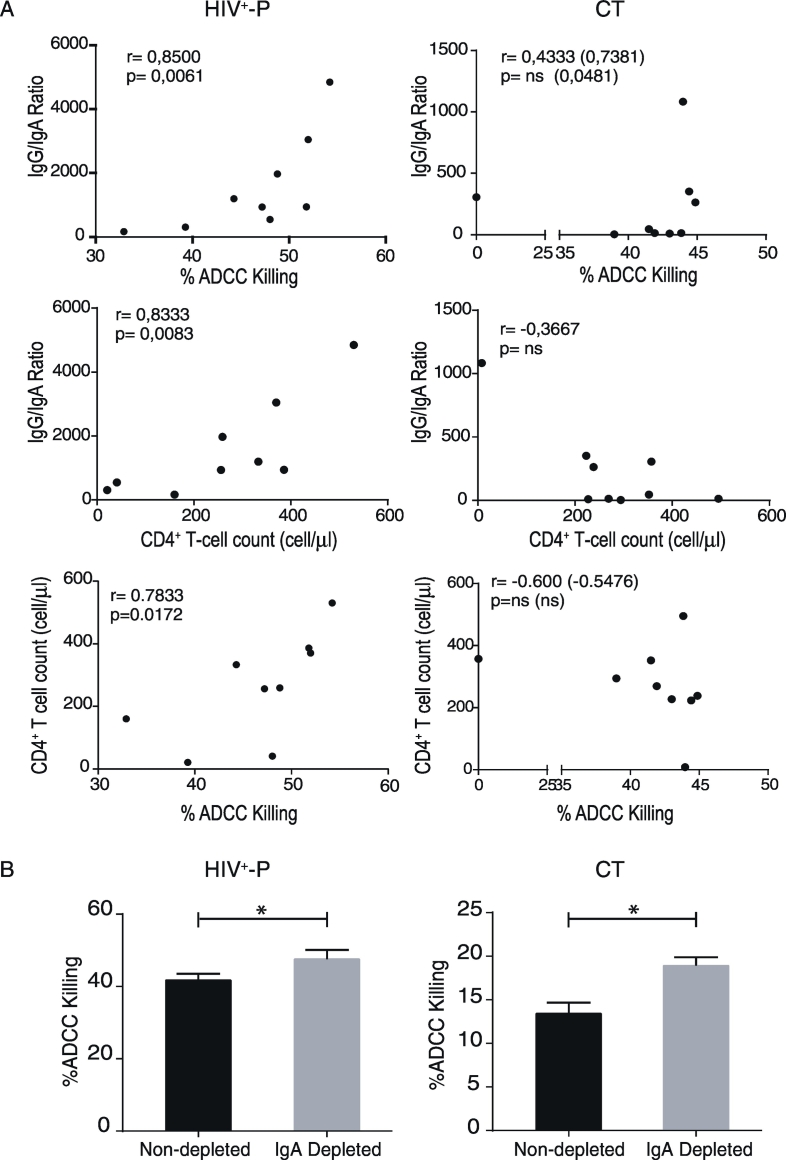
Then, we sought to evaluate immune parameters related with humoral immunity that may be implicated in resistance to HIV infection and/or related with lower transmission rates. More specifically, we aimed to determine if ADCC responses could be a novel factor involved in resistance to HIV infection. Thus, ADCC responses were initially evaluated in all HIV+-P and ESN, and in a subset of 13C and 6 HD. Zebra plots from one representative subject from each group are shown in Fig. 4A. ESN had undetectable ADCC responses, similarly to that observed in HD, excluding ADCC as a mechanism involved in protection (Fig. 4B). Interestingly, HIV+-P showed significantly higher percentages of ADCC killing (median 48.03%) compared to C (median 12.93%, p ≤ 0.0001). This result raised the hypothesis that ADCC might be involved in lowering transmission rates. However, the group of chronic subjects is a heterogeneous group that includes individuals who may potentially have transmitted the infection to others, as well as individuals who potentially may have not. So, a group of chronically infected subjects known to be HIV transmitters (CT) was specifically enrolled. By doing this, we aimed to compare levels of ADCC responses between HIV non-transmitters (HIV+-P) and documented HIV transmitters (CT). Unfortunately, limited sample was obtained from this group and only ADCC and gp120-specific antibodies could be measured in this group (no ELISPOT or plasma soluble factors). Notably, HIV+-P still showed significantly higher levels of ADCC responses (median 48.03%) compared to CT (median 43%, p = 0.039, Fig. 4B). Also, CT had significantly higher levels of ADCC compared to C (median 43% vs.12.93% respectively, p = 0.0040). When removing the outlier identified within the CT dataset, the p value between HIV+-P and CT raised to 0.07. Still, most (6 out of 9) HIV+-P had ADCC values above the IQ75% of CT. Thus, even when significance was lost after outlier removal, probably driven by the small sample size, the finding still deserves attention. Afterwards, we decided to investigate if gp120-specific IgG and IgA levels and IgG/IgA ratios may influence the ADCC responses observed in this study, as reported previously by our group in subjects undergoing primary HIV infection and ECs (Ruiz et al., 2016). For that purpose, gp120-specific IgG and IgA levels were determined. ESN had undetectable levels of both gp120-specific IgG and IgA antibodies, as in HD (Fig. 4C, left and middle panels). As expected, HIV+-P, C and CT individuals had detectable gp120-specific IgG concentrations, but no differences were observed among these groups (Fig. 4C, left panel). This was particularly interesting since HIV+-P showed higher ADCC responses both compared to C and CT, with comparable levels of anti-gp120 IgG antibodies. On the other hand, CT had significantly higher IgA titers compared to HIV+-P (p = 0.0185) and C (p = 0.0001) (Fig. 4C, middle panel). Finally, IgG/IgA ratios were calculated for the HIV-infected groups (HIV+-P, C and CT). We studied this ratio because the balance between gp120-specific IgG and IgA had been implicated in shaping ADCC responses (Tomaras et al., 2013) (Ranki et al., 1989, Cereb et al., 1995). The highest IgG/IgA ratio was observed in HIV+-P and the lowest in CT, with a statistically significant difference between them (Fig. 4C, right panel; p = 0.0040), while Cs showed an intermediate situation.
Fig. 4.
A. ADCC responses in Serodiscordant Couples (HIV+-P and ESN), HIV chronically infected individuals (C) and HIV chronically infected transmitter subjects (CT), measured by RFADCC assay. Zebra plots for one representative subject per study group. A PKH26-versus-CFSE plot was generated to determine the percentage of ADCC killing, defined as the proportion of cells that remained PKH26high but lost the viability dye (CFSEneg). Percentages shown in each plot correspond to results obtained after calculating and subtracting the background (%ADCC killing for uncoated target cells) from the media of triplicate conditions. B. Percentages of ADCC killing corresponding to a 1/1000 plasma dilution for HIV+-P, C and CT, and to a 1/100 plasma dilution for HD and ESN. %ADCC killing data correspond to background subtracted values. Horizontal lines stand for median values. Statistical analysis was performed using Kruskal-Wallis test followed by Dunn's multiple comparisons post-test. # When the outlier identified within the CT group was removed, the statistical significance was lost (p = 0.07). C. Gp120-specific IgG antibodies (log10(nanograms per milliliter), left panel), gp120-specifc IgA titers (middle panel) and IgG/IgA ratios (right panel) per study group. Horizontal lines stand for median values. Statistical analysis was performed using Kruskal-Wallis test followed by Dunn's multiple comparisons post-test. HIV+-P: HIV positive partners, ESN: Exposed HIV seronegative individuals, CT: Chronically HIV transmitters subjects C: Chronics. HD: Healthy donors.
3.5. Positive Correlations Between CD4+ T-cell Counts and Both IgG/IgA Ratios and %ADCC Killing Distinguish HIV+-P
So far, we have identified that most HIV+-P present the highest ADCC responses and that this group showed the highest IgG/IgA ratio, compared to CTs and C. Inversely, CTs showed the highest gp120-spefific IgA titers. In a previous report from our group, we showed that both the magnitude of ADCC responses and its association with disease progression were influenced by gp120-specific IgA levels (Ruiz et al., 2016). To provide a better insight into the relation of these factors in HIV+-P and CT, correlations between ADCC and gp120-specific antibodies were studied in these groups. In line with results reported previously by our group in other cohorts (Ruiz et al., 2016), gp120-specific IgG and %ADCC killing correlated positively in CT (r = 0,7857, p = 0.0279) and HIV+-P, although the correlation was not statistically significant in the latter group (r = 0.5333, p = 0.1475). On the contrary, no correlation was observed between gp120-specific IgA and %ADCC killing in either group (not shown). A strong positive correlation was found between IgG/IgA ratios and %ADCC killing in HIV+-P (r = 0.8500; p = 0.0061), suggesting that the balance between gp120-specific IgG and IgA might modulate the magnitude of ADCC responses in this group. On the other hand, this correlation was less clear in CT and only resulted statistically significant when removing the ADCC outlier data point identified within this group (Fig. 5A, upper panels). Interestingly, another unique feature of HIV+-P was a significant correlation between IgG/IgA ratios and CD4+ T-cell counts which was neither observed in CT (Fig. 5A, middle panels) nor in Chronics (r = − 0.1364, p = 0,6937, not shown).
Fig. 5.
A. Correlations between %ADCC Killing versus IgG/IgA ratios, IgG/IgA ratios versus CD4+ T-cell counts and CD4+ T-cell counts versus %ADCC killing in HIV +-P (left panels) and CT (right panels) individuals. r and p values were determined by Spearman's test. ns: non-significant. r and p values shown between brackets correspond to those obtained after the removal of the ADCC outlier data point identified within the CT group. B. Percentage of ADCC killing evaluated in IgA-depleted (grey bars) and non-depleted (black bars) plasma at 1/1000 dilutions. Plasma was obtained from representative HIV +-P and CT individuals. ADCC was evaluated in triplicates and data was analyzed by t-test.
Then, we sought to investigate if ADCC responses in HIV+-P and CTs could be related to markers of disease progression such as CD4+ T-cell count or VL. For that purpose, correlation analyses between clinical parameters and %ADCC killing observed in HIV+-P, C and CT were performed. As expected, no correlations were observed between ADCC values and VLs in either group (not shown) or CD4+ T-cell counts in Cs (r = − 0.0109, p = 0.9758 not shown) or CTs (Fig. 5A, lower panels). The lack of correlation between these parameters remained whether the outlier was included in the analysis or not. On the contrary, a significant positive correlation between %ADCC killing and CD4+ T-cell counts were observed in HIV+-P (r = 0.7833, p = 0.0172), as shown in Fig. 5A (lower panel). In contrast to our previous evidence indicating that, in viremic subjects, a protective role of ADCC antibodies was only evidenced when there were low IgA plasma levels (Ruiz et al., 2016), the scenario resulted different in HIV+-P. In this group, in spite of being viremic, ADCC response seems to play a protective role (in terms of CD4+ T-cell counts) which could also be related to lower viral transmission rates. Alternatively, it provides further evidence of the mitigating role of IgA in ADCC magnitude since this correlation in only observed in the group with the highest IgG/IgA ratio.
Based on these results, we went one step further to investigate if gp120-specific IgA from HIV+-P and CT were able to mitigate ADCC responses, as reported by our group in other cohorts (Ruiz et al., 2016). For this, plasma IgA was depleted from one representative HIV+-P and one representative CT individual. Then, ADCC was reevaluated in IgA-depleted and non-depleted plasma. As shown in fig. 5B, ADCC was modulated after IgA removal in both cases, although the extent of the modulation varied. The %ADCC killing of IgA-depleted plasma from the HIV+-P was around 13% higher compared to the undepleted plasma. However, this difference reached 33% in the CT, indicating that the IgA from the CT mitigated the ADCC responses to a greater extent than the IgA from the HIV+-P. Although these results come from only two individuals and cannot be extended to the whole groups, they provide the first evidence of IgA interference with ADCC responses in HIV+-P and CT.
Overall, these results allow us to state that, even though ADCC antibodies were associated with protection from infection in different settings (Mabuka et al., 2012, Rerks-Ngarm et al., 2009), they were not detectable in ESN. Conversely, the fact that most HIV+-P had higher percentages of ADCC killing when compared to CT made us hypothesize about an association between ADCC and a lower rate of viral transmission. This could be directly related to ADCC activity or with the balance between gp120-specific IgG and IgA antibodies. In fact, the opposite scenarios imposed by CT and HIV+-P were characterized by the highest IgA titers and the highest IgG/IgA ratios, respectively. This suggests that IgA might have a negative impact on HIV transmission. Then, ADCC levels in HIV+-P were directly associated with IgG/IgA ratios and CD4+ T-cell counts, feature uniquely found in this group. Finally, evidence of IgA-mediated mitigation of ADCC responses in HIV+-P and CT was provided. This point certainly deserves further consideration.
4. Discussion
Understanding the mechanisms underlying protection against HIV acquisition and/or disease progression will be instrumental to enhance current therapeutic regimens, as well as to develop strategies to cure HIV and to design successful vaccine candidates. In this context, the study of serodiscordant couples (SDC) gives a unique scenario to evaluate both correlates of protection in exposed HIV seronegative individuals (ESN) and also the existence of intrinsic factors in the HIV+-P that may be involved in modulating viral transmission. Among these factors, VL is a major determinant of transmission risk, i.e. reduced VL (as those found in viremic controllers) was associated with reduced transmission risk (Hull et al., 2014, Blaser et al., 2014). Within our group of HIV+-P, four out of nine subjects fell within this category thus their “viremic controller” status might be determining the lack of transmission to their partners. However, the fact that they had detectable VL and also the observation of subjects with high VLs within the HIV+-P group made us hypothesize about other factors influencing the success of the transmission event. Thus, here we assessed different aspects of the humoral and cellular HIV-specific immune responses in nine ESN and their respective HIV+-P. We covered the evaluation of (i) plasma levels of 39 cytokines and chemokines, (ii) HIV-specific T-cell responses in PBMCs, (iii) plasma gp120-specific IgG and IgA antibodies, and (iv) plasma ADCC activity. This study about different immune parameters was undertaken based on the hypothesis that resistance to HIV infection may rely on multiple host's features and also that different and combined, host, viral and environmental causes may compromise or enhance the success among transmission opportunities. Indeed, we found evidence of signatures possibly involved in diminishing transmission in HIV+-P (higher ADCC magnitude, higher IgG/IgA ratio, protective ADCC responses) versus favoring transmission efficacy in CT (higher gp120-specific IgA titers).
First, we showed that macrophage-derived chemokine (MDC) was slightly elevated in plasma from ESN, compared to HD. On the other hand, the highest plasma levels of sCD40 were found in C. Although the differences lost statistical significance after correction for multiple comparisons, a few issues are worth discussing. In previous reports, higher MDC level was associated with a better outcome in children on HAART (Lambert et al., 2007) and was directly correlated with Th17 cell counts in HIV-infected subjects (Falivene et al., 2015). Moreover, it had anti-HIV activity in in vitro experiments (Pal et al., 1997). This finding adds evidence to those reports regarding the role of innate immunity in protecting ESNs. Specifically, it has been previously reported that expression levels of the antiviral factors APOBEC3G (Biasin et al., 2007) and Mx2 (Stein et al., 2015), Vitamin D receptor (Aguilar-Jimenez et al., 2013, Aguilar-Jimenez et al., 2016), β-defensins (Zapata et al., 2008), IL-22 and acute-phase amyloid A protein (Misse et al., 2007) as well as increased responsiveness to TLR stimulation (Biasin et al., 2010) and increased NK cell activity (Ravet et al., 2007, Montoya et al., 2006, Scott-Algara et al., 2003) were associated with protection from infection in this group. Regarding sCD40L, our finding is consistent with a previous report by Sipsas et al. (2002) that showed elevated concentrations of circulating sCD40L in individuals infected with HIV-1 compared to healthy controls. Despite being infected and contrary to C, HIV+-P had sCD40L levels comparable to HD. Others have reported that sCD40L may impair dendritic cell functionality during HIV infection (Miller et al., 2015). Thus, our result would indicate that HIV+-P would be protected from this phenomenon.
Nevertheless, it is clear that the subtle differences in MDC between ESN and HD and in sCD40L between HIV+-P and C do not explain the difference behavior of each group. Thus, whether this has a direct role in HIV transmission or, more likely, other factors are also involved warrants further research. In this line, humoral and cellular HIV-specific immune responses were investigated in this study. As expected, HIV-specific cellular immune responses were broadly and readily detected by fresh ELISPOT in C and HIV+-P, and no significant differences were found between these groups. Unfortunately, ELISPOT responses could not be measured in CT due to the unavailability of PBMC samples, because this group was enrolled a posteriori and specifically for ADCC and antibodies determination. Conversely, positive responses in ESNs were infrequent (in 2 out of 6 subjects) and of very low magnitude. This result is in line with several reports indicating that detection of HIV-specific cellular immune responses in PBMCs from ESN can only be evidenced after cultured, expanded or boosted protocols and, still, subjects with positive responses are the less abundant (Ruiz-Riol et al., 2015, Guardo et al., 2015, Ritchie et al., 2011, Addo et al., 2011, Guthrie et al., 2012, Pala et al., 2013, Nguyen et al., 2006, Alimonti et al., 2006). Then, the interpretation of those positive responses should be done with caution. The most accepted hypothesis claims that it should be considered as indicators of repetitive exposure to the virus. Alternatively, these responses could be considered preformed correlates of protection against HIV infection. Out of the 2 ESN with positive ELISPOT responses in this study, one had a partner with high viral load (15,867 copies/ml) while the other one had a partner with low viral load (901 copies/ml). Thus, neither hypothesis can be excluded. This highlights that further prospective studies are needed.
Regarding humoral responses, systemic gp120-specific IgG or IgA antibodies were not detected in ESN suggesting that humoral immunity would not be involved in resistance to HIV infection. However, this hypothesis cannot be discarded since the presence of antibodies with specificities other than gp120 (for instance Gp41) as well as HIV-specific antibodies at mucosa sites should be considered. Unfortunately, we were unable to perform such evaluations for this work. In this line, one recent report indicated that ESNs may show plasma IgG or IgA that recognize cryptic Env native epitopes that cannot be detected by regular ELISA diagnostic assays (Carrillo et al., 2013). In our perspective, it is unlikely that ESN enrolled in this study had this kind of antibodies, since they would have been detected in ADCC assays (discussed below). Also, it has been demonstrated that HIV-exposed individuals that are persistently seronegative can present HIV-specific IgAs in urine and vaginal wash samples (Mazzoli et al., 1997) and also develop mucosal HIV-specific IgAs capable of blocking viral transcytosis (Lo Caputo et al., 2003). Unfortunately, we were not able to obtain this kind of samples for our study.
Finally, it has been recently shown that HIV-specific ADCC responses may generate protection from HIV infection in the context of vaccine trials (Rerks-Ngarm et al., 2009), as well as protection from disease progression in different cohorts of HIV infected subjects undergoing acute (Forthal et al., 2001, Dugast et al., 2014) and chronic infection (Lambotte et al., 2009, Thobakgale et al., 2012, Chung et al., 2011). However, there are no reports in the literature that evaluate ADCC responses in SDC. ESNs had undetectable plasma ADCC responses, similarly to that observed in HD excluding ADCC as a mechanism involved in protection. However, ADCC in other body fluids relevant to HIV transmission have not been tested so this cannot be categorically confirmed. On the other hand, HIV+-P had the highest percentages of ADCC killing. It was significantly higher than that of Cs and higher than that of most CTs. Several reports, including a report from our group, demonstrated that ECs and even VCs show enhanced ADCC function (Lambotte et al., 2009, Ackerman et al., 2016, Ruiz et al., 2016, Madhavi et al., 2017, Johansson et al., 2011). Thus, the highest ADCC observed in HIV+-P group could be influenced by the inclusion of VCs within this group. To exclude this possibility, ADCC was compared between VC HIV+-P versus non-VC HIV+-P and no significant difference was found (data not shown, p = 0,68). This reinforced the hypothesis about the potential role of ADCC modulating HIV transmission. This result is particularly relevant given that similar findings have been reported in other settings but never in a scenario involving sexual transmission. For instance, Mabuka and colleagues (Mabuka et al., 2012) demonstrated that mother-to-child transmission decreases due to the presence of antibodies capable of mediating ADCC present in breast milk, associating these antibodies with lower transmission rates. Another recent study provided evidence indicating that pre-existing levels of IgG1 type antibodies in infants born to HIV+ mothers correlated with increased ADCC responses and lower mortality upon infection (Milligan et al., 2015). Recently, Parsons et al. (Parsons et al., 2016) reported that seminal plasma from HIV infected individuals on HAART contained antibodies capable of mediating ADCC. These findings add to existing data accounting for the presence of antibodies capable of mediating ADCC in vaginal and cervical fluids (Battle-Miller et al., 2002, Nag et al., 2004). Taken together, these evidences provide rational criteria for investigating the presence of ADCC-mediating antibodies in mucosa tissue and body fluids from HIV+ individuals engaged in SDCs and their potential role on HIV transmission. It is worth noting here that, even when the most relevant result found in this study is the fact that HIV+-P showed significantly higher ADCC than C, and that most HIV+-P showed higher ADCC than CT, the observation that both HIV+-P and CT revealed significantly higher percentages of ADCC killing compared to C cannot be overlooked. The magnitude of ADCC response found in C was similar to that observed in a previous study from our group for a similar cohort of chronically infected subjects (Ruiz et al., 2016), confirming this result. Alternatively, these results may suggest that ADCC and IgA might have a differential impact on HIV transmission. In this line, higher ADCC levels observed in HIV+-P were associated with higher IgG/IgA ratios or, in other words, a more favourable balance between the levels of gp120-specific IgG and IgA. On the other hand, CTs showed the lowest IgG/IgA ratios and the highest levels of gp120-specific IgA. Thus, high ADCC responses but in a context of elevated IgA might be predisposing to transmission. Moreover, it was found that gp120-specific IgA compromised ADCC responses in both HIV+-P and CT with different intensities, although this remains to be confirmed in subsequent studies. This finding adds evidence to previous results from our group demonstrating that gp120-specific IgA was capable of modifying ADCC responses, not only in the vaccine setting as demonstrated by Tomaras et al. (Tomaras et al., 2013) but also during progressive natural HIV infection (Ruiz et al., 2016).
New to this study is the fact that significant correlations between CD4+ T-cell count versus IgG/IgA ratios and versus %ADCC killing were only found in HIV+-P allowing us to identify signatures that define non-transmitter HIV+ individuals. The fact that these positive correlations were only seen in individuals with the highest IgG/IgA ratios (HIV+-P) is in consonance with our previous observations found in viremic subjects, where a protective role of ADCC antibodies is only evidenced after plasma IgA removal (Ruiz et al., 2016). Besides providing further support to this notion, these results allow to postulate that these factors (ADCC and IgG/IgA balance) could be related to lower viral transmission rates. It remains to be determined whether antibody specificities, avidity, Ig subclasses or any other factors are involved in this effect. It is also of interest to uncover the reasons underlying the differential magnitude of gp120-specific IgA between HIV+-P and CTs.
One point that raises concern when analyzing HIV-specific immune responses is viral variability. Responses can be severely underestimated if not choosing the proper reagents. Particularly in Argentina, B and BF subtypes are the most prevalent, followed by a minor proportion of subtype C infections (Dilernia et al., 2013). The frequency of viral variants found in this work matches those observed at a population level (Table 1). In this scenario, cellular responses were evaluated using the multiclade PTE peptide panels. Our group have previously used these panels in samples obtained from BF infected subjects from Argentina (Turk et al., 2013, Ghiglione et al., 2014, Rodriguez et al., 2012) proving its efficiency. Regarding humoral responses, the gp120 protein used in the ADCC and in the ELISA assays to detect gp120-specific IgG and IgA was a recombinant gp120 derived from a subtype B strain. Hypothetically, this could lead to an underestimation of the response in non-B infected subjects. Unfortunately, no recombinant gp120 protein from CRF12_BF strains or other BF recombinant forms are available so as to use the protein corresponding to the autologous subtype in the assays. To overcome this issue, we compared %ADCC killing and HIV-specific IgG and IgA levels between B and BF infected subjects from our study groups. No differences were observed when segregating the subjects according to the viral subtype, across all study groups (data not shown). Thus, at least in this study, results are not being influenced at this level.
Broadly neutralizing antibodies are able to inhibit HIV cell-to-cell transmission as described by Malbec et al. (Malbec et al., 2013). But if this is also the case of ADCC antibodies, still needs to be proved. Therefore, it would be particularly interesting to examine whether ADCC serum and mucosal antibodies may also contribute to avoid HIV-1 cell-to-cell spread in vitro, and the way this mechanism is modulated by IgG and IgA levels.
To the best of our knowledge, this is the first report evaluating multiple factors potentially involved in HIV transmission and in protection from acquiring the infection in a SDC cohort from Argentina. More importantly, this is the first report that evaluates ADCC responses in the context of HIV SDC and in comparison with C and CT. We found possible factors involved in modulating transmission rates. Particularly, it was shown that HIV+-P had higher ADCC levels, most likely driven by a beneficial IgG/IgA balance. However, CTs showed the highest gp120-specific IgA levels. Correlation analyses showed that ADCC responses and IgG/IgA balance were directly associated with CD4+ T-cell counts uniquely in HIV+-P. Also, initial evidence is provided regarding a mitigating role of gp120-specific IgA on ADCC responses in HIV+-P and CT. These data provide key aspects to understand the host factors involved in this phenomenon and allow the visualization of further investigations. Moreover, it also provides support for mechanisms that should be considered both in the development of prophylactic vaccines as well as novel immunotherapies against HIV infection.
Funding Sources
This work was supported by grants from the Agencia Nacional de Promoción Científica y Tecnológica and GlaxoSmithKline (PICT2012, Grant # 0475 and PICTO-GSK, Grant # 2013/0006) and from Universidad de Buenos Aires (UBACyT 2013–2016, Grant # 20020120200263BA) to GT. This investigation also used resources that were supported by the Southwest National Primate Research Center grant P51 OD011133 from the Office of Research Infrastructure Programs, National Institutes of Health (NIH) to LDG. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Conflict of Interest
The authors have no conflicts of interest to report.
Authors Contributions
M.J.R., M.M.G and G.T. conceived and designed the experiments; M.J.R., J.S., Y.G, C.C, and C.T. performed the experiments; M.J.R., G.T, Y.G, A.M.R, L.D.G., N.L., H.S, and M.M.G analyzed the data; L.A, G.D, L.D.G, and O·S contributed samples/analysis tools; M.J.R. and G.T wrote the paper.
Acknowledgments
Acknowledgments
Authors specially acknowledge study participants for agreeing to collaborate in this study and to provide blood samples. CCR5Δ32 typing was gently performed by Dr. Andrea Mangano at the Laboratorio de Biología Celular y Retrovirus, CONICET, Hospital de Pediatría “Prof. Dr. Juan P. Garrahan”, Buenos Aires, Argentina. We thank Mr. Sergio Mazzini for language assistance during manuscript preparation and Dr. Cecilia Quiroga (Instituto de Microbiología (IMPaM), UBA, Buenos Aires, Argentina) for assitance with statistical analysis. Authors have no conflict of interests to declare.
References
- Ackerman M.E., Mikhailova A., Brown E.P., Dowell K.G., Walker B.D., Bailey-Kellogg C., Suscovich T.J., Alter G. Polyfunctional HIV-specific antibody responses are associated with spontaneous HIV control. PLoS Pathog. 2016;12 doi: 10.1371/journal.ppat.1005315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Addo M.M., Altfeld M., Brainard D.M., Rathod A., Piechocka-Trocha A., Fideli U., Mulenga J., Shutes E., Alvino D.M., Hunter E., Allen S.A., Walker B.D. Lack of detectable HIV-1-specific CD8(+) T cell responses in Zambian HIV-1-exposed seronegative partners of HIV-1-positive individuals. J. Infect. Dis. 2011;203:258–262. doi: 10.1093/infdis/jiq028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilar-Jimenez W., Zapata W., Caruz A., Rugeles M.T. High transcript levels of vitamin D receptor are correlated with higher mRNA expression of human beta defensins and IL-10 in mucosa of HIV-1-exposed seronegative individuals. PLoS One. 2013;8 doi: 10.1371/journal.pone.0082717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilar-Jimenez W., Zapata W., Rugeles M.T. Antiviral molecules correlate with vitamin D pathway genes and are associated with natural resistance to HIV-1 infection. Microbes Infect. 2016;18:510–516. doi: 10.1016/j.micinf.2016.03.015. [DOI] [PubMed] [Google Scholar]
- Alimonti J.B., Kimani J., Matu L., Wachihi C., Kaul R., Plummer F.A., Fowke K.R. Characterization of CD8 T-cell responses in HIV-1-exposed seronegative commercial sex workers from Nairobi, Kenya. Immunol. Cell Biol. 2006;84:482–485. doi: 10.1111/j.1440-1711.2006.01455.x. [DOI] [PubMed] [Google Scholar]
- Battle-Miller K., Eby C.A., Landay A.L., Cohen M.H., Sha B.E., Baum L.L. Antibody-dependent cell-mediated cytotoxicity in cervical lavage fluids of human immunodeficiency virus type 1—infected women. J. Infect. Dis. 2002;185:439–447. doi: 10.1086/338828. [DOI] [PubMed] [Google Scholar]
- Baum L.L., Cassutt K.J., Knigge K., Khattri R., Margolick J., Rinaldo C., Kleeberger C.A., Nishanian P., Henrard D.R., Phair J. HIV-1 gp120-specific antibody-dependent cell-mediated cytotoxicity correlates with rate of disease progression. J. Immunol. 1996;157:2168–2173. [PubMed] [Google Scholar]
- Biasin M., Piacentini L., Lo Caputo S., Kanari Y., Magri G., Trabattoni D., Naddeo V., Lopalco L., Clivio A., Cesana E., Fasano F., Bergamaschi C., Mazzotta F., Miyazawa M., Clerici M. Apolipoprotein B mRNA-editing enzyme, catalytic polypeptide-like 3G: a possible role in the resistance to HIV of HIV-exposed seronegative individuals. J. Infect. Dis. 2007;195:960–964. doi: 10.1086/511988. [DOI] [PubMed] [Google Scholar]
- Biasin M., Piacentini L., Lo Caputo S., Naddeo V., Pierotti P., Borelli M., Trabattoni D., Mazzotta F., Shearer G.M., Clerici M. TLR activation pathways in HIV-1-exposed seronegative individuals. J. Immunol. 2010;184:2710–2717. doi: 10.4049/jimmunol.0902463. [DOI] [PubMed] [Google Scholar]
- Blaser N., Wettstein C., Estill J., Vizcaya L.S., Wandeler G., Egger M., Keiser O. Impact of viral load and the duration of primary infection on HIV transmission: systematic review and meta-analysis. AIDS. 2014;28:1021–1029. doi: 10.1097/QAD.0000000000000135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrillo J., Restrepo C., Rallon N.I., Massanella M., del Romero J., Rodriguez C., Soriano V., Clotet B., Benito J.M., Blanco J. HIV exposed seronegative individuals show antibodies specifically recognizing native HIV envelope glycoprotein. AIDS. 2013;27:1375–1385. doi: 10.1097/QAD.0b013e32835fac08. [DOI] [PubMed] [Google Scholar]
- Cereb N., Maye P., Lee S., Kong Y., Yang S.Y. Locus-specific amplification of HLA class I genes from genomic DNA: locus-specific sequences in the first and third introns of HLA-A, -B, and -C alleles. Tissue Antigens. 1995;45:1–11. doi: 10.1111/j.1399-0039.1995.tb02408.x. [DOI] [PubMed] [Google Scholar]
- Cheynier R., Langlade-Demoyen P., Marescot M.R., Blanche S., Blondin G., Wain-Hobson S., Griscelli C., Vilmer E., Plata F. Cytotoxic T lymphocyte responses in the peripheral blood of children born to human immunodeficiency virus-1-infected mothers. Eur. J. Immunol. 1992;22:2211–2217. doi: 10.1002/eji.1830220905. [DOI] [PubMed] [Google Scholar]
- Chung A.W., Navis M., Isitman G., Wren L., Silvers J., Amin J., Kent S.J., Stratov I. Activation of NK cells by ADCC antibodies and HIV disease progression. J. Acquir. Immune Defic. Syndr. 2011;58:127–131. doi: 10.1097/QAI.0b013e31822c62b9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clerici M., Levin J.M., Kessler H.A., Harris A., Berzofsky J.A., Landay A.L., Shearer G.M. HIV-specific T-helper activity in seronegative health care workers exposed to contaminated blood. JAMA. 1994;271:42–46. [PubMed] [Google Scholar]
- Coloccini R.S., Dilernia D., Ghiglione Y., Turk G., Laufer N., Rubio A., Socias M.E., Figueroa M.I., Sued O., Cahn P., Salomon H., Mangano A., Pando M.A. Host genetic factors associated with symptomatic primary HIV infection and disease progression among Argentinean seroconverters. PLoS One. 2014;9 doi: 10.1371/journal.pone.0113146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Currier J.R., Kuta E.G., Turk E., Earhart L.B., Loomis-Price L., Janetzki S., Ferrari G., Birx D.L., Cox J.H. A panel of MHC class I restricted viral peptides for use as a quality control for vaccine trial ELISPOT assays. J. Immunol. Methods. 2002;260:157–172. doi: 10.1016/s0022-1759(01)00535-x. [DOI] [PubMed] [Google Scholar]
- Dale B.M., Alvarez R.A., Chen B.K. Mechanisms of enhanced HIV spread through T-cell virological synapses. Immunol. Rev. 2013;251:113–124. doi: 10.1111/imr.12022. [DOI] [PubMed] [Google Scholar]
- Deeks S.G., Lewin S.R., Ross A.L., Ananworanich J., Benkirane M., Cannon P., Chomont N., Douek D., Lifson J.D., Lo Y.R., Kuritzkes D., Margolis D., Mellors J., Persaud D., Tucker J.D., Barre-Sinoussi F., International ASTaCWG, Alter G., Auerbach J., Autran B., Barouch D.H., Behrens G., Cavazzana M., Chen Z., Cohen E.A., Corbelli G.M., Eholie S., Eyal N., Fidler S., Garcia L., Grossman C., Henderson G., Henrich T.J., Jefferys R., Kiem H.P., McCune J., Moodley K., Newman P.A., Nijhuis M., Nsubuga M.S., Ott M., Palmer S., Richman D., Saez-Cirion A., Sharp M., Siliciano J., Silvestri G., Singh J., Spire B., Taylor J., Tolstrup M., Valente S., van Lunzen J., Walensky R., Wilson I., Zack J. International AIDS society global scientific strategy: towards an HIV cure 2016. Nat. Med. 2016;22:839–850. doi: 10.1038/nm.4108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dilernia D.A., Monaco D.C., Cesar C., Krolewiecki A.J., Friedman S.R., Cahn P., Salomon H. Estimation of HIV-testing rates to maximize early diagnosis-derived benefits at the individual and population level. PLoS One. 2013;8 doi: 10.1371/journal.pone.0053193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dugast A.S., Stamatatos L., Tonelli A., Suscovich T.J., Licht A.F., Mikell I., Ackerman M.E., Streeck H., Klasse P.J., Moore J.P., Alter G. Independent evolution of Fc- and Fab-mediated HIV-1-specific antiviral antibody activity following acute infection. Eur. J. Immunol. 2014;44:2925–2937. doi: 10.1002/eji.201344305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falivene J., Ghiglione Y., Laufer N., Socias M.E., Holgado M.P., Ruiz M.J., Maeto C., Figueroa M.I., Giavedoni L.D., Cahn P., Salomon H., Sued O., Turk G., Gherardi M.M. Th17 and Th17/Treg ratio at early HIV infection associate with protective HIV-specific CD8(+) T-cell responses and disease progression. Sci. Rep. 2015;5 doi: 10.1038/srep11511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Vina M.A., Lazaro A.M., Marcos C.Y., Nulf C., Raimondi E., Haas E.J., Stastny P. Dissimilar evolution of B-locus versus A-locus and class II loci of the HLA region in South American Indian tribes. Tissue Antigens. 1997;50:233–250. doi: 10.1111/j.1399-0039.1997.tb02867.x. [DOI] [PubMed] [Google Scholar]
- Forthal D.N., Landucci G., Daar E.S. Antibody from patients with acute human immunodeficiency virus (HIV) infection inhibits primary strains of HIV type 1 in the presence of natural-killer effector cells. J. Virol. 2001;75:6953–6961. doi: 10.1128/JVI.75.15.6953-6961.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowke K.R., Nagelkerke N.J., Kimani J., Simonsen J.N., Anzala A.O., Bwayo J.J., MacDonald K.S., Ngugi E.N., Plummer F.A. Resistance to HIV-1 infection among persistently seronegative prostitutes in Nairobi, Kenya. Lancet. 1996;348:1347–1351. doi: 10.1016/S0140-6736(95)12269-2. [DOI] [PubMed] [Google Scholar]
- Frank E.H., Witten I.H. The WEKA workbench. In: Kaufmann M., editor. Data Mining: Practical Machine Learning Tools and Techniques. 2016. [Google Scholar]
- Ghiglione Y., Falivene J., Ruiz M.J., Laufer N., Socias M.E., Cahn P., Giavedoni L., Sued O., Gherardi M.M., Salomon H., Turk G. Early skewed distribution of total and HIV-specific CD8 + T-cell memory phenotypes during primary HIV infection is related to reduced antiviral activity and faster disease progression. PLoS One. 2014;9:e104235. doi: 10.1371/journal.pone.0104235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giavedoni L.D. Simultaneous detection of multiple cytokines and chemokines from nonhuman primates using luminex technology. J. Immunol. Methods. 2005;301:89–101. doi: 10.1016/j.jim.2005.03.015. [DOI] [PubMed] [Google Scholar]
- Gomez-Roman V.R., Florese R.H., Patterson L.J., Peng B., Venzon D., Aldrich K., Robert-Guroff M. A simplified method for the rapid fluorometric assessment of antibody-dependent cell-mediated cytotoxicity. J. Immunol. Methods. 2006;308:53–67. doi: 10.1016/j.jim.2005.09.018. [DOI] [PubMed] [Google Scholar]
- Gonzalo-Gil E., Ikediobi U., Sutton R.E. Mechanisms of virologic control and clinical characteristics of HIV + elite/viremic controllers. Yale J. Biol. Med. 2017;90:245–259. [PMC free article] [PubMed] [Google Scholar]
- Goonetilleke N., Moore S., Dally L., Winstone N., Cebere I., Mahmoud A., Pinheiro S., Gillespie G., Brown D., Loach V., Roberts J., Guimaraes-Walker A., Hayes P., Loughran K., Smith C., De Bont J., Verlinde C., Vooijs D., Schmidt C., Boaz M., Gilmour J., Fast P., Dorrell L., Hanke T., McMichael A.J. Induction of multifunctional human immunodeficiency virus type 1 (HIV-1)-specific T cells capable of proliferation in healthy subjects by using a prime-boost regimen of DNA- and modified vaccinia virus Ankara-vectored vaccines expressing HIV-1 Gag coupled to CD8 + T-cell epitopes. J. Virol. 2006;80:4717–4728. doi: 10.1128/JVI.80.10.4717-4728.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guardo A.C., Ruiz-Riol M., Fernandez E., Maleno M.J., Bargallo M.E., Leon A., Climent N., Garcia F., Gatell J.M., Brander C., Plana M. Detection of HIV-1-specific T-cell immune responses in highly HIV-exposed uninfected individuals by in-vitro dendritic cell co-culture. AIDS. 2015;29:1309–1318. doi: 10.1097/QAD.0000000000000728. [DOI] [PubMed] [Google Scholar]
- Guthrie B.L., Lohman-Payne B., Liu A.Y., Bosire R., Nuvor S.V., Choi R.Y., Mackelprang R.D., Kiarie J.N., De Rosa S.C., Richardson B.A., John-Stewart G.C., Farquhar C. HIV-1-specific enzyme-linked immunosorbent spot assay responses in HIV-1-exposed uninfected partners in discordant relationships compared to those in low-risk controls. Clin. Vaccine Immunol. 2012;19:1798–1805. doi: 10.1128/CVI.00179-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hladik F., McElrath M.J. Setting the stage: host invasion by HIV. Nat. Rev. Immunol. 2008;8:447–457. doi: 10.1038/nri2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell D.N., Andreotti P.E., Dawson J.R., Cresswell P. Natural killing target antigens as inducers of interferon: studies with an immunoselected, natural killing-resistant human T lymphoblastoid cell line. J. Immunol. 1985;134:971–976. [PubMed] [Google Scholar]
- Hull M., Lange J., Montaner J.S. Treatment as prevention—where next? Curr. HIV/AIDS Rep. 2014;11:496–504. doi: 10.1007/s11904-014-0237-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson S.E., Rollman E., Chung A.W., Center R.J., Hejdeman B., Stratov I., Hinkula J., Wahren B., Karre K., Kent S.J., Berg L. NK cell function and antibodies mediating ADCC in HIV-1-infected viremic and controller patients. Viral Immunol. 2011;24:359–368. doi: 10.1089/vim.2011.0025. [DOI] [PubMed] [Google Scholar]
- Katoh K., Standley D.M. MAFFT: iterative refinement and additional methods. Methods Mol. Biol. 2014;1079:131–146. doi: 10.1007/978-1-62703-646-7_8. [DOI] [PubMed] [Google Scholar]
- Lambert J.S., Machado E.S., Watson D.C., Sill A.M., Lim J.K., Charurat M., Cunha S.M., Afonso A.O., Oliviera R.H., Tanuri A., DeVico A.L. Production of the HIV-suppressive chemokines CCL3/MIP-1alpha and CCL22/MDC is associated with more effective antiretroviral therapy in HIV-infected children. Pediatr. Infect. Dis. J. 2007;26:935–944. doi: 10.1097/INF.0b013e31812714db. [DOI] [PubMed] [Google Scholar]
- Lambotte O., Ferrari G., Moog C., Yates N.L., Liao H.X., Parks R.J., Hicks C.B., Owzar K., Tomaras G.D., Montefiori D.C., Haynes B.F., Delfraissy J.F. Heterogeneous neutralizing antibody and antibody-dependent cell cytotoxicity responses in HIV-1 elite controllers. AIDS. 2009;23:897–906. doi: 10.1097/QAD.0b013e328329f97d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langlade-Demoyen P., Ngo-Giang-Huong N., Ferchal F., Oksenhendler E. Human immunodeficiency virus (HIV) nef-specific cytotoxic T lymphocytes in noninfected heterosexual contact of HIV-infected patients. J. Clin. Invest. 1994;93:1293–1297. doi: 10.1172/JCI117085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R., Paxton W.A., Choe S., Ceradini D., Martin S.R., Horuk R., MacDonald M.E., Stuhlmann H., Koup R.A., Landau N.R. Homozygous defect in HIV-1 coreceptor accounts for resistance of some multiply-exposed individuals to HIV-1 infection. Cell. 1996;86:367–377. doi: 10.1016/s0092-8674(00)80110-5. [DOI] [PubMed] [Google Scholar]
- Lo Caputo S., Trabattoni D., Vichi F., Piconi S., Lopalco L., Villa M.L., Mazzotta F., Clerici M. Mucosal and systemic HIV-1-specific immunity in HIV-1-exposed but uninfected heterosexual men. AIDS. 2003;17:531–539. doi: 10.1097/00002030-200303070-00008. [DOI] [PubMed] [Google Scholar]
- Mabuka J., Nduati R., Odem-Davis K., Peterson D., Overbaugh J. HIV-specific antibodies capable of ADCC are common in breastmilk and are associated with reduced risk of transmission in women with high viral loads. PLoS Pathog. 2012;8:e1002739. doi: 10.1371/journal.ppat.1002739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhavi V., Wines B.D., Amin J., Emery S., Group ES, Lopez E., Kelleher A., Sydney L.S.G., Center R.J., Hogarth P.M., Chung A.W., Kent S.J., Stratov I. HIV-1 Env- and Vpu-specific antibody-dependent cellular cytotoxicity responses associated with elite control of HIV. J. Virol. 2017:91. doi: 10.1128/JVI.00700-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malbec M., Porrot F., Rua R., Horwitz J., Klein F., Halper-Stromberg A., Scheid J.F., Eden C., Mouquet H., Nussenzweig M.C., Schwartz O. Broadly neutralizing antibodies that inhibit HIV-1 cell to cell transmission. J. Exp. Med. 2013;210:2813–2821. doi: 10.1084/jem.20131244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malhotra U., Li F., Nolin J., Allison M., Zhao H., Mullins J.I., Self S., McElrath M.J. Enhanced detection of human immunodeficiency virus type 1 (HIV-1) Nef-specific T cells recognizing multiple variants in early HIV-1 infection. J. Virol. 2007;81:5225–5237. doi: 10.1128/JVI.02564-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin D.P., Murrell B., Golden M., Khoosal A., Muhire B. RDP4: detection and analysis of recombination patterns in virus genomes. Virus Evol. 2015;1 doi: 10.1093/ve/vev003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maydt J., Lengauer T. Recco: recombination analysis using cost optimization. Bioinformatics. 2006;22:1064–1071. doi: 10.1093/bioinformatics/btl057. [DOI] [PubMed] [Google Scholar]
- Mazzoli S., Trabattoni D., Lo Caputo S., Piconi S., Ble C., Meacci F., Ruzzante S., Salvi A., Semplici F., Longhi R., Fusi M.L., Tofani N., Biasin M., Villa M.L., Mazzotta F., Clerici M. HIV-specific mucosal and cellular immunity in HIV-seronegative partners of HIV-seropositive individuals. Nat. Med. 1997;3:1250–1257. doi: 10.1038/nm1197-1250. [DOI] [PubMed] [Google Scholar]
- McDermott A.B., Koup R.A. CD8(+) T cells in preventing HIV infection and disease. AIDS. 2012;26:1281–1292. doi: 10.1097/QAD.0b013e328353bcaf. [DOI] [PubMed] [Google Scholar]
- Miller E.A., Gopal R., Valdes V., Berger J.S., Bhardwaj N., O'Brien M.P. Soluble CD40 ligand contributes to dendritic cell-mediated T-cell dysfunction in HIV-1 infection. AIDS. 2015;29:1287–1296. doi: 10.1097/QAD.0000000000000698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milligan C., Richardson B.A., John-Stewart G., Nduati R., Overbaugh J. Passively acquired antibody-dependent cellular cytotoxicity (ADCC) activity in HIV-infected infants is associated with reduced mortality. Cell Host Microbe. 2015;17:500–506. doi: 10.1016/j.chom.2015.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misse D., Yssel H., Trabattoni D., Oblet C., Lo Caputo S., Mazzotta F., Pene J., Gonzalez J.P., Clerici M., Veas F. IL-22 participates in an innate anti-HIV-1 host-resistance network through acute-phase protein induction. J. Immunol. 2007;178:407–415. doi: 10.4049/jimmunol.178.1.407. [DOI] [PubMed] [Google Scholar]
- Montoya C.J., Velilla P.A., Chougnet C., Landay A.L., Rugeles M.T. Increased IFN-gamma production by NK and CD3 +/CD56 + cells in sexually HIV-1-exposed but uninfected individuals. Clin. Immunol. 2006;120:138–146. doi: 10.1016/j.clim.2006.02.008. [DOI] [PubMed] [Google Scholar]
- Nag P., Kim J., Sapiega V., Landay A.L., Bremer J.W., Mestecky J., Reichelderfer P., Kovacs A., Cohn J., Weiser B., Baum L.L. Women with cervicovaginal antibody-dependent cell-mediated cytotoxicity have lower genital HIV-1 RNA loads. J. Infect. Dis. 2004;190:1970–1978. doi: 10.1086/425582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen M., Pean P., Lopalco L., Nouhin J., Phoung V., Ly N., Vermisse P., Henin Y., Barre-Sinoussi F., Burastero S.E., Reynes J.M., Carcelain G., Pancino G. HIV-specific antibodies but not t-cell responses are associated with protection in seronegative partners of HIV-1-infected individuals in Cambodia. J. Acquir. Immune Defic. Syndr. 2006;42:412–419. doi: 10.1097/01.qai.0000222289.97825.35. [DOI] [PubMed] [Google Scholar]
- Pal R., Garzino-Demo A., Markham P.D., Burns J., Brown M., Gallo R.C., DeVico A.L. Inhibition of HIV-1 infection by the beta-chemokine MDC. Science. 1997;278:695–698. doi: 10.1126/science.278.5338.695. [DOI] [PubMed] [Google Scholar]
- Pala P., Serwanga J., Watera C., Ritchie A.J., Moodie Z., Wang M., Goonetilleke N., Birabwa E., Hughes P., Senkaali D., Nakiboneka R., Grosskurth H., Haynes B., McMichael A., Kaleebu P., Center for HIVAVI Quantitative and qualitative differences in the T cell response to HIV in uninfected Ugandans exposed or unexposed to HIV-infected partners. J. Virol. 2013;87:9053–9063. doi: 10.1128/JVI.00721-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons M.S., Madhavi V., Ana-Sosa-Batiz F., Center R.J., Wilson K.M., Bunupuradah T., Ruxrungtham K., Kent S.J. Brief report: seminal plasma anti-HIV antibodies trigger antibody-dependent cellular cytotoxicity: implications for HIV transmission. J. Acquir. Immune Defic. Syndr. 2016;71:17–23. doi: 10.1097/QAI.0000000000000804. [DOI] [PubMed] [Google Scholar]
- Piacentini L., Fenizia C., Naddeo V., Clerici M. Not just sheer luck! Immune correlates of protection against HIV-1 infection. Vaccine. 2008;26:3002–3007. doi: 10.1016/j.vaccine.2007.11.062. [DOI] [PubMed] [Google Scholar]
- Pinto L.A., Sullivan J., Berzofsky J.A., Clerici M., Kessler H.A., Landay A.L., Shearer G.M. ENV-specific cytotoxic T lymphocyte responses in HIV seronegative health care workers occupationally exposed to HIV-contaminated body fluids. J. Clin. Invest. 1995;96:867–876. doi: 10.1172/JCI118133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranki A., Mattinen S., Yarchoan R., Broder S., Ghrayeb J., Lahdevirta J., Krohn K. T-cell response towards HIV in infected individuals with and without zidovudine therapy, and in HIV-exposed sexual partners. AIDS. 1989;3:63–69. doi: 10.1097/00002030-198902000-00002. [DOI] [PubMed] [Google Scholar]
- Ravet S., Scott-Algara D., Bonnet E., Tran H.K., Tran T., Nguyen N., Truong L.X., Theodorou I., Barre-Sinoussi F., Pancino G., Paul P. Distinctive NK-cell receptor repertoires sustain high-level constitutive NK-cell activation in HIV-exposed uninfected individuals. Blood. 2007;109:4296–4305. doi: 10.1182/blood-2006-08-040238. [DOI] [PubMed] [Google Scholar]
- Rerks-Ngarm S., Pitisuttithum P., Nitayaphan S., Kaewkungwal J., Chiu J., Paris R., Premsri N., Namwat C., de Souza M., Adams E., Benenson M., Gurunathan S., Tartaglia J., McNeil J.G., Francis D.P., Stablein D., Birx D.L., Chunsuttiwat S., Khamboonruang C., Thongcharoen P., Robb M.L., Michael N.L., Kunasol P., Kim J.H., Investigators M.-T. Vaccination with ALVAC and AIDSVAX to prevent HIV-1 infection in Thailand. N. Engl. J. Med. 2009;361:2209–2220. doi: 10.1056/NEJMoa0908492. [DOI] [PubMed] [Google Scholar]
- Ritchie A.J., Campion S.L., Kopycinski J., Moodie Z., Wang Z.M., Pandya K., Moore S., Liu M.K., Brackenridge S., Kuldanek K., Legg K., Cohen M.S., Delwart E.L., Haynes B.F., Fidler S., McMichael A.J., Goonetilleke N. Differences in HIV-specific T cell responses between HIV-exposed and -unexposed HIV-seronegative individuals. J. Virol. 2011;85:3507–3516. doi: 10.1128/JVI.02444-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez A.M., Turk G., Pascutti M.F., Ferrer F., Najera J.L., Monaco D., Esteban M., Salomon H., Calamante G., Gherardi M.M. Characterization of DNA and MVA vectors expressing Nef from HIV-1 CRF12_BF revealed high immune specificity with low cross-reactivity against subtype B. Virus Res. 2009;146:1–12. doi: 10.1016/j.virusres.2009.08.004. [DOI] [PubMed] [Google Scholar]
- Rodriguez A.M., Pascutti M.F., Maeto C., Falivene J., Holgado M.P., Turk G., Gherardi M.M. IL-12 and GM-CSF in DNA/MVA immunizations against HIV-1 CRF12_BF Nef induced T-cell responses with an enhanced magnitude, breadth and quality. PLoS One. 2012;7:e37801. doi: 10.1371/journal.pone.0037801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowland-Jones S.L., McMichael A. Immune responses in HIV-exposed seronegatives: have they repelled the virus? Curr. Opin. Immunol. 1995;7:448–455. doi: 10.1016/0952-7915(95)80087-5. [DOI] [PubMed] [Google Scholar]
- Rowland-Jones S.L., Nixon D.F., Aldhous M.C., Gotch F., Ariyoshi K., Hallam N., Kroll J.S., Froebel K., McMichael A. HIV-specific cytotoxic T-cell activity in an HIV-exposed but uninfected infant. Lancet. 1993;341:860–861. doi: 10.1016/0140-6736(93)93063-7. [DOI] [PubMed] [Google Scholar]
- Rowland-Jones S., Sutton J., Ariyoshi K., Dong T., Gotch F., McAdam S., Whitby D., Sabally S., Gallimore A., Corrah T. HIV-specific cytotoxic T-cells in HIV-exposed but uninfected Gambian women. Nat. Med. 1995;1:59–64. doi: 10.1038/nm0195-59. [DOI] [PubMed] [Google Scholar]
- Ruiz M.J., Ghiglione Y., Falivene J., Laufer N., Holgado M.P., Socias M.E., Cahn P., Sued O., Giavedoni L., Salomon H., Gherardi M.M., Rodriguez A.M., Turk G. Env-specific IgA from viremic HIV-infected subjects compromises antibody-dependent cellular cytotoxicity. J. Virol. 2016;90:670–681. doi: 10.1128/JVI.02363-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Riol M., Llano A., Ibarrondo J., Zamarreno J., Yusim K., Bach V., Mothe B., Perez-Alvarez S., Fernandez M.A., Requena G., Meulbroek M., Pujol F., Leon A., Cobarsi P., Korber B.T., Clotet B., Ganoza C., Sanchez J., Coll J., Brander C. Alternative effector-function profiling identifies broad HIV-specific T-cell responses in highly HIV-exposed individuals who remain uninfected. J. Infect. Dis. 2015;211:936–946. doi: 10.1093/infdis/jiu534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz A.K., Zhang M., Bulla I., Leitner T., Korber B., Morgenstern B., Stanke M. jpHMM: improving the reliability of recombination prediction in HIV-1. Nucleic Acids Res. 2009;37:W647–651. doi: 10.1093/nar/gkp371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott-Algara D., Truong L.X., Versmisse P., David A., Luong T.T., Nguyen N.V., Theodorou I., Barre-Sinoussi F., Pancino G. Cutting edge: increased NK cell activity in HIV-1-exposed but uninfected Vietnamese intravascular drug users. J. Immunol. 2003;171:5663–5667. doi: 10.4049/jimmunol.171.11.5663. [DOI] [PubMed] [Google Scholar]
- Sipsas N.V., Sfikakis P.P., Kontos A., Kordossis T. Levels of soluble CD40 ligand (CD154) in serum are increased in human immunodeficiency virus type 1-infected patients and correlate with CD4(+) T-cell counts. Clin. Diagn. Lab. Immunol. 2002;9:558–561. doi: 10.1128/CDLI.9.3.558-561.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein D.R., Shaw S.Y., McKinnon L.R., Abou M., McCorrister S.J., Westmacott G.R., Fowke K.R., Plummer F.A., Ball T.B. Mx2 expression is associated with reduced susceptibility to HIV infection in highly exposed HIV seronegative Kenyan sex workers. AIDS. 2015;29:35–41. doi: 10.1097/QAD.0000000000000490. [DOI] [PubMed] [Google Scholar]
- Thobakgale C.F., Fadda L., Lane K., Toth I., Pereyra F., Bazner S., Ndung'u T., Walker B.D., Rosenberg E.S., Alter G., Carrington M., Allen T.M., Altfeld M. Frequent and strong antibody-mediated natural killer cell activation in response to HIV-1 Env in individuals with chronic HIV-1 infection. J. Virol. 2012;86:6986–6993. doi: 10.1128/JVI.00569-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomaras G.D., Ferrari G., Shen X., Alam S.M., Liao H.X., Pollara J., Bonsignori M., Moody M.A., Fong Y., Chen X., Poling B., Nicholson C.O., Zhang R., Lu X., Parks R., Kaewkungwal J., Nitayaphan S., Pitisuttithum P., Rerks-Ngarm S., Gilbert P.B., Kim J.H., Michael N.L., Montefiori D.C., Haynes B.F. Vaccine-induced plasma IgA specific for the C1 region of the HIV-1 envelope blocks binding and effector function of IgG. Proc. Natl. Acad. Sci. U. S. A. 2013;110:9019–9024. doi: 10.1073/pnas.1301456110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turk G., Gherardi M.M., Laufer N., Saracco M., Luzzi R., Cox J.H., Cahn P., Salomon H. Magnitude, breadth, and functional profile of T-cell responses during human immunodeficiency virus primary infection with B and BF viral variants. J. Virol. 2008;82:2853–2866. doi: 10.1128/JVI.02260-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turk G., Ghiglione Y., Falivene J., Socias M.E., Laufer N., Coloccini R.S., Rodriguez A.M., Ruiz M.J., Pando M.A., Giavedoni L.D., Cahn P., Sued O., Salomon H., Gherardi M.M. Early Gag immunodominance of the HIV-specific T-cell response during acute/early infection is associated with higher CD8 + T-cell antiviral activity and correlates with preservation of the CD4 + T-cell compartment. J. Virol. 2013;87:7445–7462. doi: 10.1128/JVI.00865-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zapata W., Rodriguez B., Weber J., Estrada H., Quinones-Mateu M.E., Zimermman P.A., Lederman M.M., Rugeles M.T. Increased levels of human beta-defensins mRNA in sexually HIV-1 exposed but uninfected individuals. Curr. HIV Res. 2008;6:531–538. doi: 10.2174/157016208786501463. [DOI] [PMC free article] [PubMed] [Google Scholar]