Fig. 6.
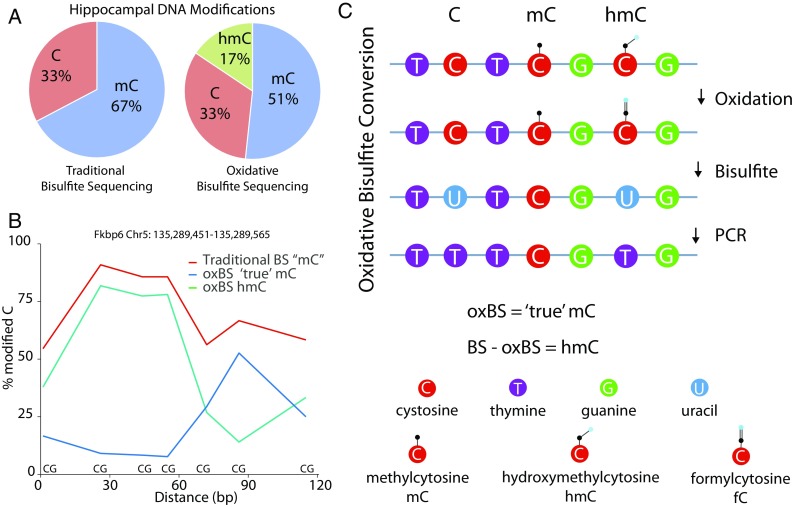
Hydroxymethylation analysis. Most DNA modification analysis studies use bisulfite sequencing approaches that do not differentiate between methylation and hydroxymethylation. However, hydroxymethylation is abundant in a number of tissues. For example, using data from Hadad et al. (2016) who examined the mouse hippocampus by oxidative bisulfite sequencing that differentiates between methylation and hydroxymethylation, it is clear (a) that one fourth of what would normally be called as methylation is in fact hydroxymethylation. Further demonstrating this point, within a relatively small region (~ 120 bp of Fkbp6; b), the levels of methylation and hydroxymethylation are clearly regulated in a base-specific fashion. Hydroxymethylation can be differentiated from methylation by an oxidation step (c) that de-protects hydroxymethylation and allows a specific methylation quantitation. When combined with bisulfite sequencing, a subtractive approach can be used to quantify methylation and hydroxymethylation individually
