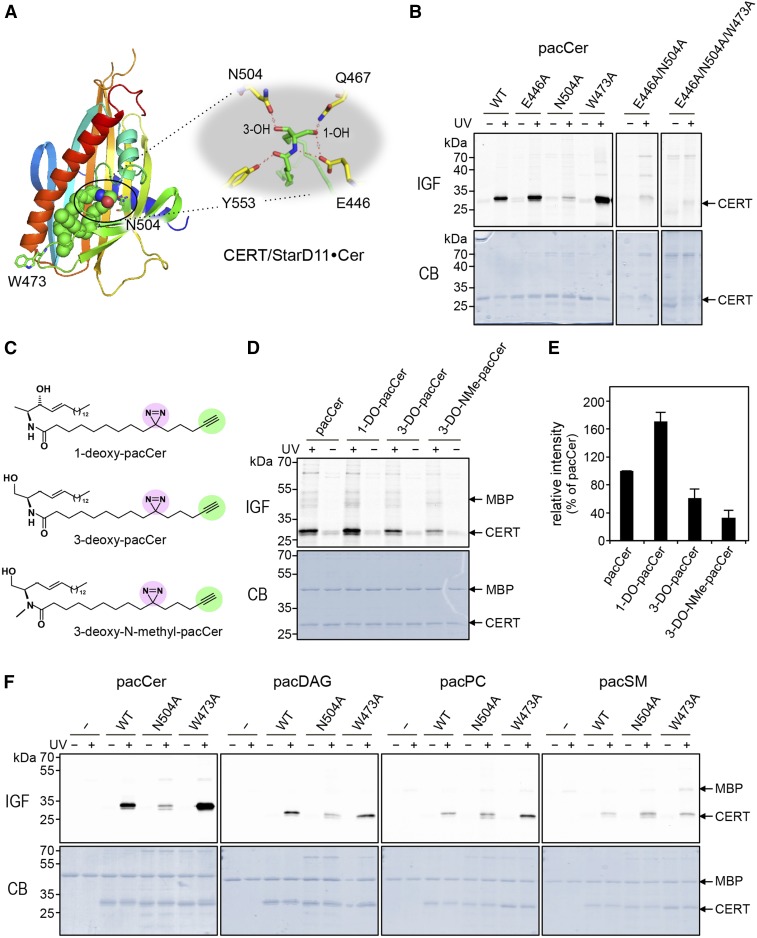
Fig. 2.
Probing the lipid-binding pocket of CERT using bifunctional lipid analogs. A: Structure of the START domain of CERT in complex with ceramide (PDB 2E3O). The START domain is displayed as a rainbow-colored ribbon representation and the ceramide molecule as spheres, with green, red, and blue color representing C, O, and N atoms, respectively. The small inset highlights residues involved in coordinating the ceramide molecule in the lipid-binding pocket. B: WT and point-mutants of CERT START were produced in E. coli, purified, and then incubated with liposomes containing 1 mol% of pacCer for 30 min at 37°C. Samples were either UV-irradiated (+) or kept in the dark (−), clickreacted with Alexa Fluor647-N3, and then analyzed by in-gel-fluorescence (IGF) and Coomassie staining (CB). C: Bifunctional analogs of C1-deoxy-ceramide (1-DO-pacCer), C3-deoxy-ceramide (3-DO-pacCer), and C3-deoxy-N-methyl-ceramide (3-DO-NMe-pacCer). D: WT CERT START was mixed with maltose-binding protein (MBP), incubated with liposomes containing 1 mol% of pacCer, 1-DO-pacCer, 3-DO-pacCer, or 3-DO-NMe-pacCer, and processed as in B. E: Quantitative analysis of photoaffinity labeling of CERT START by deoxy derivatives of pacCer relative to control (pacCer). The labeling intensity of CERT START by pacCer was set at 100%. Data are shown as the mean ± SD of three independent experiments. F: WT and point-mutants of CERT START were mixed with MBP, incubated with liposomes containing 1 mol% of pacCer, pacDAG, pacPC, or pacSM, and processed as in B.