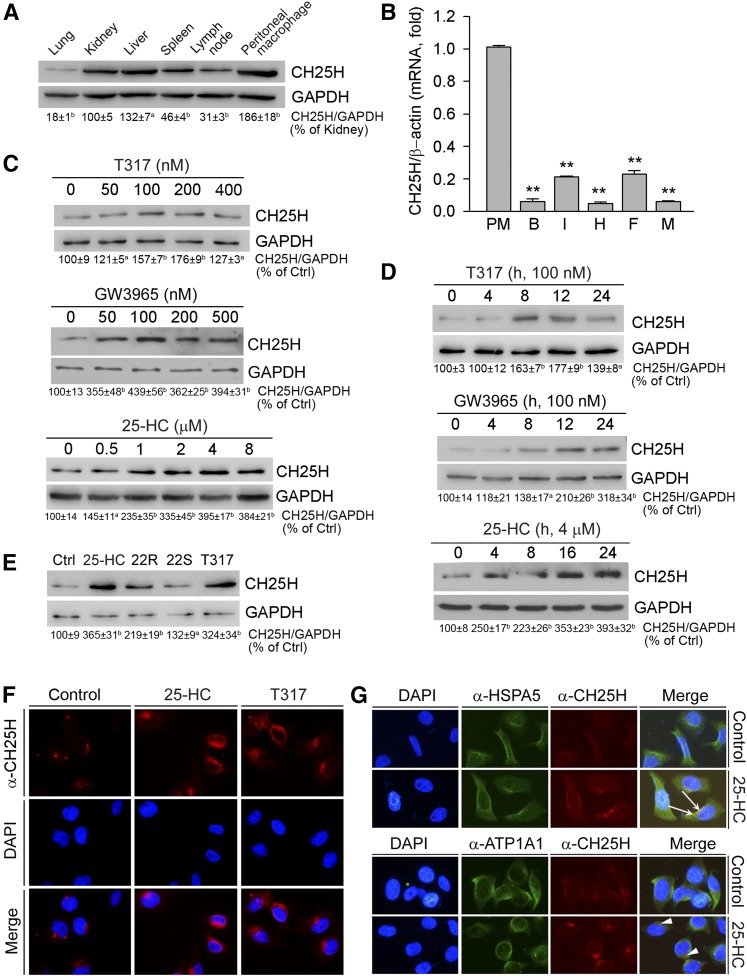
Fig. 1.
Activation of LXR induces hepatic CH25H protein expression. A: Total cellular proteins were extracted from C57BL/6 mouse lung, kidney, liver, spleen, lymph node, and peritoneal macrophages. B: Total RNA was extracted from mouse peritoneal macrophages (PM), brain (B), intestine (I), heart (H), adipose tissue (F) and muscle (M). C: HepG2 cells in serum-free medium were treated with T317, GW3965, or 25-HC for 16 h. D: HepG2 cells were treated with T317 (100 nM), GW3965 (100 nM), or 25-HC (4 μM) for the indicated times. E: HepG2 cells were treated with 25-HC (2 μM), 22(R)-hydroxycholesterol (22R, 2 μM), 22(S)-hydroxycholesterol (22S, 2 μM), or T317 (200 nM) for 16 h. Expression of CH25H protein (A, C–E) was determined by Western blot. The bands from three repeated experiments were scanned and the density was quantified with statistical analysis; aP < 0.05; bP < 0.01 vs. control (n = 3). Expression of CH25H mRNA (B) was determined by quantitative real-time-PCR; **P < 0.01 vs. peritoneal macrophages (n = 4). F, G: HepG2 cells were treated with 25-HC (2 μM) or T317 (200 nM) for 16 h. Expression of CH25H protein (F, G), or HSPA5 and ATP1A1 protein (G) was determined by immunofluorescent staining. The white arrows indicate the colocalization of CH25H and HSPA5 proteins; the arrowheads indicate the different localization of CH25H protein from the plasma membrane.