Abstract
During various surveys of Phytophthora diversity in Europe, Chile and Vietnam slow growing oomycete isolates were obtained from rhizosphere soil samples and small streams in natural and planted forest stands. Phylogenetic analyses of sequences from the nuclear ITS, LSU, β-tubulin and HSP90 loci and the mitochondrial cox1 and NADH1 genes revealed they belong to six new species of a new genus, officially described here as Nothophytophthora gen. nov., which clustered as sister group to Phytophthora. Nothophytophthora species share numerous morphological characters with Phytophthora: persistent (all Nothophytophthora spp.) and caducous (N. caduca, N. chlamydospora, N. valdiviana, N. vietnamensis) sporangia with variable shapes, internal differentiation of zoospores and internal, nested and extended (N. caduca, N. chlamydospora) and external (all Nothophytophthora spp.) sporangial proliferation; smooth-walled oogonia with amphigynous (N. amphigynosa) and paragynous (N. amphigynosa, N. intricata, N. vietnamensis) attachment of the antheridia; chlamydospores (N. chlamydospora) and hyphal swellings. Main differing features of the new genus are the presence of a conspicuous, opaque plug inside the sporangiophore close to the base of most mature sporangia in all known Nothophytophthora species and intraspecific co-occurrence of caducity and non-papillate sporangia with internal nested and extended proliferation in several Nothophytophthora species. Comparisons of morphological structures of both genera allow hypotheses about the morphology and ecology of their common ancestor which are discussed. Production of caducous sporangia by N. caduca, N. chlamydospora and N. valdiviana from Valdivian rainforests and N. vietnamensis from a mountain forest in Vietnam suggests a partially aerial lifestyle as adaptation to these humid habitats. Presence of tree dieback in all forests from which Nothophytophthora spp. were recovered and partial sporangial caducity of several Nothophytophthora species indicate a pathogenic rather than a saprophytic lifestyle. Isolation tests from symptomatic plant tissues in these forests and pathogenicity tests are urgently required to clarify the lifestyle of the six Nothophytophthora species.
Keywords: breeding system, caducity, evolution, oomycetes, Peronosporaceae, phylogeny
INTRODUCTION
The Peronosporaceae, a sister family of the Pythiaceae, belongs to the Peronosporales, class Peronosporomycetes, kingdom Stramenipila, and currently comprises 22 genera, i.e., Phytophthora, Halophytophthora, Phytopythium and 19 genera of downy mildews (Dick 2001, Hulvey et al. 2010, Beakes et al. 2014, Thines & Choi 2016). While Halophytophthora and Phytopythium species are mostly saprophytes and/or necrotrophic facultative plant pathogens most Phytophthora species have a hemibiotrophic or necrotrophic lifestyle as primary plant pathogens although for mostly aquatic Phytophthora species a partially saprophytic lifestyle seems likely (Erwin & Ribeiro 1996, Brasier et al. 2003, Jung et al. 2011). In contrast, all c. 600 species of downy mildews are highly specialized, obligate biotrophic plant pathogens (Göker et al. 2007, Runge et al. 2011, Beakes et al. 2012, Thines & Choi 2016). However, the production of RxLR-type effectors, which play a crucial role for pathogenesis, by both Phytophthora and the downy mildews indicates a close relationship between the two groups (Baxter et al. 2010, Thines & Kamoun 2010). Several phylogenetic studies demonstrated that the genus Phytophthora is monophyletic and that all downy mildews reside within Phytophthora (Cooke et al. 2000, Kroon et al. 2004, Göker et al. 2007, Runge et al. 2011, Martin et al. 2014, Thines & Choi 2016). However, due to the description of the obligate biotrophic downy mildews as 19 distinct genera, mainly before the advent of molecular phylogenetic analyses, Phytophthora exhibits a high degree of paraphyly (Cooke et al. 2000, Göker et al. 2007, Runge et al. 2011, Thines & Choi 2016). The molecular results confirmed the hypothesis of Gäumann (1952) who, based on morphological and pathogenic data, postulated an evolutionary development from saprophytic Pythium species via hemibiotrophic or necrotrophic Phytophthora species to the obligate biotrophic downy mildews. Unlike Phytophthora, the genus Pythium was in DNA sequence-based phylogenetic analyses shown to be polyphyletic (Briard et al. 1995, Cooke et al. 2000, De Cock & Lévesque 2004, Kroon et al. 2004, Villa et al. 2006). Consequently, the genus was recently divided in Pythium s.str. and four new genera, i.e., Phytopythium (syn. Ovatisporangium; previously Pythium Clade K), Elongisporangium, Globisporangium and Pilasporangium (Bala et al. 2010, Uzuhashi et al. 2010, De Cock et al. 2015). While Phytopythium together with the other four genera was originally assigned to the Pythiaceae (De Cock et al. 2015), Thines & Choi (2016) considered Phytopythium belonging to the Peronosporaceae due to both phylogenetic relatedness and morphological similarity to Phytophthora.
Stimulated by the increasing number of epidemics caused by exotic invasive Phytophthora species including P. austrocedri, P. cinnamomi, P. lateralis, P. plurivora, P. ramorum, P. xalni or P. xcambivora in both managed and natural ecosystems (Brasier et al. 1993, Erwin & Ribeiro 1996, Jung et al. 1996, 2000, 2013, 2016, Hansen et al. 2000, 2012, Rizzo et al. 2002, Vettraino et al. 2002, 2005, Balci & Halmschlager 2003a, b, 2007, Jung & Blaschke 2004, Hardham 2005, Greslebin et al. 2007, Jung 2009, Jung & Burgess 2009, Brasier & Webber 2010, Green et al. 2013, 2015, Ginetti et al. 2014, Henricot et al. 2014, Scanu et al. 2015) numerous Phytophthora surveys have been performed during the past two decades in forests and river systems in most continents. Using classical isolation methods and, more recently, also metagenomic approaches, these surveys have uncovered an astonishing diversity of described and previously unknown Phytophthora taxa (Jung et al. 2000, 2011, 2013, 2016, 2017a, b, Balci & Halmschlager 2003a, b, 2007, Jung 2009, Zeng et al. 2009, Rea et al. 2011, Reeser et al. 2011, Hansen et al. 2012, Huai et al. 2013, Hüberli et al. 2013, Oh et al. 2013, Shrestha et al. 2013, Burgess 2015, Català et al. 2015, Burgess et al. 2017).
During surveys of Phytophthora diversity in Europe, Chile and Vietnam slow growing isolates which morphologically resemble Phytophthora species were obtained from rhizosphere soil samples and small streams in natural and planted forest stands. A preliminary phylogenetic analysis of ITS rDNA sequences resulted in six distinct clades belonging to a potentially new genus in sister position with Phytophthora. In this study, morphological and physiological characteristics were used in combination with DNA sequence data from four nuclear gene regions, i.e., ITS, part of the 28S large subunit (LSU), heat shock protein 90 (HSP90) and β-tubulin (Btub), and the two mitochondrial cox1 and NADH1 genes to characterise and officially describe the new oomycete genus as Nothophytophthora gen. nov., and the six new taxa as N. amphigynosa sp. nov., N. caduca sp. nov., N. chlamydospora sp. nov., N. intricata sp. nov., N. valdiviana sp. nov. and N. vietnamensis sp. nov.
MATERIAL AND METHODS
Isolate collection and maintenance
Details of all isolates used in the phylogenetic, morphological and temperature-growth studies are given in Table 1. Sampling and isolation methods from forest soil and streams were according to Jung et al. (1996, 2017a). For baiting of soils young leaves of Lithocarpus bacgiangensis (Vietnam), and Fagus sylvatica and Quercus robur (Germany) were used as baits. Stream baiting was performed using young leaves of Castanea sativa, F. sylvatica, Nothofagus obliqua and Q. robur (Chile), and Citrus sinensis and Quercus suber (Portugal). For all isolates, single hyphal tip cultures were produced under the stereomicroscope from the margins of fresh cultures on V8-juice agar (V8A; 16 g agar, 3 g CaCO3, 100 mL Campbell’s V8 juice, 900 mL distilled water). Stock cultures were maintained on grated carrot agar (CA; 16 g agar, 3 g CaCO3, 200 g carrots, 1 000 mL distilled water; Brasier 1967, Scanu et al. 2015) at 10 °C in the dark. All isolates of the six new Nothophytophthora spp. are preserved in the culture collections maintained at the University of Algarve, the University of Sassari and the Plant Protection Institute, Centre for Agricultural Research, Hungarian Academy of Sciences. Ex-type and isotype cultures were deposited at the Westerdijk Fungal Biodiversity Institute (previously Centraalbureau voor Schimmelcultures CBS; Utrecht, The Netherlands) (Table 1).
Table 1.
Details of isolates from Nothophytophthora and related genera considered in the phylogenetic, morphological and growth-temperature studies. GenBank numbers for sequences obtained in the present study are printed in italics.
| Species |
Isolate numbersa |
Origin |
GenBank accession numbers |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| International collections | Local collections | Host; source | Location; year | Collector; reference | ITS | LSU | Btub | HSP90 | Cox1 | NADH1 | |
| N. amphigynosabcd | CBS 142348 | BD268 | Stream baiting; atlantic forest | Portugal; 2015 | T. Jung; this study | KY788382 | KY788428 | KY788515 | KY788555 | KY788473 | KY788596 |
| N. amphigynosabcd | – | BD269 | Stream baiting; atlantic forest | Portugal; 2015 | T. Jung; this study | KY788383 | KY788431 | KY788516 | KY788556 | KY788474 | KY788597 |
| N. amphigynosabcd | CBS 142349 | BD741 | Stream baiting; atlantic forest | Portugal; 2015 | T. Jung; this study | KY788384 | KY788432 | KY788517 | KY788557 | KY788475 | KY788598 |
| N. amphigynosabcd | – | BD742 | Stream baiting; atlantic forest | Portugal; 2015 | T. Jung; this study | KY788385 | KY788434 | KY788518 | KY788558 | KY788476 | KY788599 |
| N. amphigynosabc | – | BD857 | Stream baiting; atlantic forest | Portugal; 2015 | T. Jung; this study | KY788386 | KY788429 | KY788519 | KY788559 | KY788477 | KY788600 |
| N. amphigynosabc | – | BD858 | Stream baiting; atlantic forest | Portugal; 2015 | T. Jung; this study | KY788387 | KY788430 | KY788520 | KY788560 | KY788478 | KY788601 |
| N. amphigynosabc | – | BD859 | Stream baiting; atlantic forest | Portugal; 2015 | T. Jung; this study | KY788388 | KY788435 | KY788521 | KY788561 | KY788479 | KY788602 |
| N. amphigynosabc | – | BD860 | Stream baiting; atlantic forest | Portugal; 2015 | T. Jung; this study | KY788389 | KY788433 | KY788522 | KY788562 | KY788480 | KY788603 |
| N. caducabcd | CBS 142350 | CL328 | Stream baiting; Valdivian rainforest | Chile; 2014 | T. Jung; this study | KY788401 | KY788470 | KY788531 | KY788571 | KY788489 | KY788612 |
| N. caducacd | – | CL235b | Stream baiting; Valdivian rainforest | Chile; 2014 | T. Jung; this study | KY788390 | KY788459 | – | – | – | – |
| N. caducabc | – | CL239 | Stream baiting; Valdivian rainforest | Chile; 2014 | T. Jung; this study | KY788391 | KY788460 | KY788523 | KY788563 | KY788481 | KY788604 |
| N. caducabc | – | CL240 | Stream baiting; Valdivian rainforest | Chile; 2014 | T. Jung; this study | KY788392 | KY788461 | KY788524 | KY788564 | KY788482 | KY788605 |
| N. caducabcd | – | CL320 | Stream baiting; Valdivian rainforest | Chile; 2014 | T. Jung; this study | KY788393 | KY788462 | KY788525 | KY788565 | KY788483 | KY788606 |
| N. caducabcd | – | CL321 | Stream baiting; Valdivian rainforest | Chile; 2014 | T. Jung; this study | KY788394 | KY788463 | KY788526 | KY788566 | KY788484 | KY788607 |
| N. caducabcd | – | CL322 | Stream baiting; Valdivian rainforest | Chile; 2014 | T. Jung; this study | KY788395 | KY788464 | – | – | – | – |
| N. caducabcd | – | CL323 | Stream baiting; Valdivian rainforest | Chile; 2014 | T. Jung; this study | KY788396 | KY788465 | KY788527 | KY788567 | KY788485 | KY788608 |
| N. caducabcd | – | CL324 | Stream baiting; Valdivian rainforest | Chile; 2014 | T. Jung; this study | KY788397 | KY788466 | – | – | – | – |
| N. caducabcd | – | CL325 | Stream baiting; Valdivian rainforest | Chile; 2014 | T. Jung; this study | KY788398 | KY788467 | KY788528 | KY788568 | KY788486 | KY788609 |
| N. caducabcd | – | CL326 | Stream baiting; Valdivian rainforest | Chile; 2014 | T. Jung; this study | KY788399 | KY788468 | KY788529 | KY788569 | KY788487 | KY788610 |
| N. caducabcd | – | CL327 | Stream baiting; Valdivian rainforest | Chile; 2014 | T. Jung; this study | KY788400 | KY788469 | KY788530 | KY788570 | KY788488 | KY788611 |
| N. caducabc | CBS 142351 | CL333 | Stream baiting; Valdivian rainforest | Chile; 2014 | T. Jung; this study | KY788402 | KY788471 | KY788532 | KY788572 | KY788490 | KY788613 |
| N. caducabc | – | CL334 | Stream baiting; Valdivian rainforest | Chile; 2014 | T. Jung; this study | KY788403 | KY788472 | KY788533 | KY788573 | KY788491 | KY788614 |
| N. chlamydosporabcd | CBS 142353 | CL316 | Stream baiting; Valdivian rainforest | Chile; 2014 | T. Jung; this study | KY788405 | KY788450 | KY788535 | KY788575 | KY788493 | KY788616 |
| N. chlamydosporabc | CBS 142352 | CL195 | Stream baiting; Valdivian rainforest | Chile; 2014 | T. Jung; this study | KY788404 | KY788449 | KY788534 | KY788574 | KY788492 | KY788615 |
| N. chlamydosporabcd | – | CL317 | Stream baiting; Valdivian rainforest | Chile; 2014 | T. Jung; this study | KY788406 | KY788451 | KY788536 | KY788576 | KY788494 | KY788617 |
| N. chlamydosporabcd | – | CL318 | Stream baiting; Valdivian rainforest | Chile; 2014 | T. Jung; this study | KY788407 | KY788452 | KY788537 | KY788577 | KY788495 | KY788618 |
| N. chlamydosporabcd | – | CL319 | Stream baiting; Valdivian rainforest | Chile; 2014 | T. Jung; this study | KY788408 | KY788453 | KY788538 | KY788578 | KY788496 | KY788619 |
| N. intricatabcd | CBS 142354 | RK113-1s | Aesculus hippocastanum | Germany; 2011 | T. Jung; this study | KY788413 | KY788440 | KY788543 | KY788583 | KY788501 | KY788624 |
| N. intricatac | – | RK113-1sa | A. hippocastanum | Germany; 2011 | T. Jung; this study | – | – | – | – | – | – |
| N. intricatabcd | – | RK113-1sb | A. hippocastanum | Germany; 2011 | T. Jung; this study | KY788409 | KY788436 | KY788539 | KY788579 | KY788497 | KY788620 |
| N. intricatabcd | CBS 142355 | RK113-1sH | A. hippocastanum | Germany; 2011 | T. Jung; this study | KY788412 | KY788439 | KY788542 | KY788582 | KY788500 | KY788623 |
| N. intricatabcd | – | RK113-1sHa | A. hippocastanum | Germany; 2011 | T. Jung; this study | KY788410 | KY788437 | KY788540 | KY788580 | KY788498 | KY788621 |
| N. intricatabcd | – | RK113-1sHb | A. hippocastanum | Germany; 2011 | T. Jung; this study | KY788411 | KY788438 | KY788541 | KY788581 | KY788499 | KY788622 |
| N. valdivianabcd | CBS 142357 | CL331 | Stream baiting; Valdivian rainforest | Chile; 2014 | T. Jung; this study | KY788417 | KY788457 | KY788547 | KY788587 | KY788505 | KY788628 |
| N. valdivianabc | CBS 142356 | CL242 | Stream baiting; Valdivian rainforest | Chile; 2014 | T. Jung; this study | KY788414 | KY788454 | KY788544 | KY788584 | KY788502 | KY788625 |
| N. valdivianabcd | – | CL329 | Stream baiting; Valdivian rainforest | Chile; 2014 | T. Jung; this study | KY788415 | KY788455 | KY788545 | KY788585 | KY788503 | KY788626 |
| N. valdivianabcd | – | CL330 | Stream baiting; Valdivian rainforest | Chile; 2014 | T. Jung; this study | KY788416 | KY788456 | KY788546 | KY788586 | KY788504 | KY788627 |
| N. valdivianabcd | – | CL332 | Stream baiting; Valdivian rainforest | Chile; 2014 | T. Jung; this study | KY788418 | KY788458 | KY788548 | KY788588 | KY788506 | KY788629 |
| N. vietnamensisbcd | CBS 142358 | VN794 | Castanopsis sp. & Acer campbellii | Vietnam; 2016 | T. Jung; this study | KY788420 | KY788442 | KY788550 | KY788590 | KY788508 | KY788631 |
| N. vietnamensisbcd | – | VN230 | Castanopsis sp. & Acer campbellii | Vietnam; 2016 | T. Jung; this study | KY788419 | KY788441 | KY788549 | KY788589 | KY788507 | KY788630 |
| N. vietnamensisbcd | CBS 142359 | VN795 | Castanopsis sp. & Acer campbellii | Vietnam; 2016 | T. Jung; this study | KY788421 | KY788443 | KY788551 | KY788591 | KY788509 | KY788632 |
| N. vietnamensiscd | – | VN796 | Castanopsis sp. & Acer campbellii | Vietnam; 2016 | T. Jung; this study | KY788422 | KY788444 | – | – | – | – |
| N. vietnamensiscd | – | VN797 | Castanopsis sp. & Acer campbellii | Vietnam; 2016 | T. Jung; this study | KY788423 | KY788445 | – | – | – | – |
| N. vietnamensiscd | – | VN798 | Castanopsis sp. & Acer campbellii | Vietnam; 2016 | T. Jung; this study | KY788424 | KY788446 | – | – | – | – |
| N. vietnamensisbcd | – | VN799 | Castanopsis sp. & Acer campbellii | Vietnam; 2016 | T. Jung; this study | KY788425 | KY788447 | KY788552 | KY788592 | KY788510 | KY788633 |
| N. vietnamensisbcd | – | VN800 | Castanopsis sp. & Acer campbellii | Vietnam; 2016 | T. Jung; this study | KY788426 | KY788448 | KY788553 | KY788593 | KY788511 | KY788634 |
| Nothophytophthora sp.be | – | REB326-69 | Stream baiting | New Zealand; 2008 | –; Than et al. 2013 | JX122744 | – | – | – | – | – |
| Nothophytophthora sp.bf | – | PR12-475 | Stream baiting | Ireland; 2014 | –; O’Hanlon et al. 2016 | KT633937 | – | – | – | – | – |
| Nothophytophthora sp.bg | – | PR13-109 | Stream baiting | Ireland; 2015 | –; O’Hanlon et al. 2016 | KT633938 | – | – | – | – | – |
| Aphanomyces euteichesb | CBS 156.73 | ||||||||||
| IMI 170485 | – | Pisum sativum | Norway; – | L. Sundheim; Robideau et al. 2011 | HQ643117 | HQ665132 | – | – | HQ708190 | – | |
| Elongisporangium anandrumb | CBS 285.31 | – | Rheum rhaponticum | – | C. Drechsler; Robideau et al. 2011 | HQ643435 | HQ665185 | – | – | HQ708482 | – |
| E. undulatumb | CBS 157.69 | ||||||||||
| IMI 323158 | – | Soil under Pinus sp. | Alabama; 1968 | W.A. Campbell; Robideau et al. 2011 | HQ643946 | HQ665134 | – | – | HQ708987 | – | |
| Halophytophthora avicenniaeb | CBS 188.85 | ||||||||||
| ATCC 64709 | DAR 50187 | Avicennia marina | Australia; – | S. Wilkens; Robideau et al. 2011 | HQ643147 | HQ665146 | – | – | HQ708219 | – | |
| H. batemanensisb | CBS 679.84 | ||||||||||
| IMI 327602 | DAR 41559 | Soil-covered leaf of Avicennia sp. | Australia; 1982 | J. Simpson; Robideau et al. 2011 | HQ643148 | HQ665286 | – | – | HQ708220 | – | |
| H. epistomiab | CBS 590.85 | ||||||||||
| ATCC 28293 | |||||||||||
| IMI 330183 | – | Decaying leaf | Florida; – | I.M. Master & J.W. Fell; Robideau et al. 2011 | HQ643220 | HQ665279 | – | – | HQ708285 | – | |
| H. exoproliferab | CBS 252.93 | ||||||||||
| ATCC 76607 | IFO 32420 | Fallen leaf of Bruguiera gymnorrhyza | Japan (Okinawa island); 1988 | –; Robideau et al. 2011 | HQ643132 | HQ665174 | – | – | HQ708205 | – | |
| H. operculatab | CBS 241.83 | ||||||||||
| ATCC 44952 | – | Decaying leaf of Avicennia marina | Australia, – | –; De Cock et al. 2015 | KJ128038 | KJ128038 | – | – | KF853238 | – | |
| H. polymorphicab | CBS 680.84 | DAR 41562 | Eucalyptus sp. | Australia; 1982 | J. Simpson; Robideau et al. 2011, De Cock et al. 2015 | HQ643313 | HQ665288 | – | – | HQ708363 | – |
| Hyaloperonospora sisymbrii-sophiaeb | HV276 | – | Descurainia sophia | Austria; 2000 | H. Voglmayr; Voglmayr 2003 | AY198253 | EU054910 | – | – | HM033186 | – |
| Peronospora rumicisb | HV312 | – | Rumex acetosa | Austria; 2000 | H. Voglmayr; Voglmayr 2003 | AY198287 | KC495032 | – | – | KC494952 | – |
| Phytophthora asparagib | WPC P10690 | ||||||||||
| ICMP 9495 | – | Asparagus officinalis | New Zealand; 1986 | P.G. Falloon; Robideau et al. 2011 | HQ261683 | EU080569 | – | – | HQ261430 | – | |
| P. boehmeriaeb | CBS 291.29 | ||||||||||
| IMI 180614 | – | Boehmeria nivea | Japan; – | K. Sawada; Robideau et al. 2011 | HQ643149 | HQ665190 | EU080162 | EU080165 | HQ708221 | AY563992 | |
| P. cactorumb | WPC P0714 | – | Syringa vulgaris | The Netherlands; 1930 | W.L. White; Robideau et al. 2011 | HQ261514 | EU080282 | – | – | HQ261261 | – |
| P. captiosab | WPC P10719 | ||||||||||
| ICMP 15576 | – | Eucalyptus saligna | New Zealand; 1992 | M.A. Dick & C.W. Barr; Robideau et al. 2011 | HQ261522 | EU079663 | – | – | HQ261269 | – | |
| P. castaneaeb | CBS 587.85 | ||||||||||
| ATCC 36818 | |||||||||||
| IMI 325914 | – | Soil | Taiwan; – | H.S. Chang; Robideau et al. 2011 | HQ643255 | HQ665278 | – | – | HQ708315 | – | |
| P. colocasiaeb | WPC P6317 | – | Colocasia esculenta | Indonesia; 1989 | M.D. Coffey; Robideau et al. 2011 | HQ261539 | EU079911 | – | – | HQ261286 | – |
| P. foliorumb | WPC P10969 | 1307997-MI | Rhododendron sp. | California; 2005 | C. Blomquist; Robideau et al. 2011 | HQ261561 | EU079684 | – | – | HQ261308 | – |
| P. heveaeb | CBS 296.29 | ||||||||||
| IMI 180616 | – | Hevea brasiliensis | Malaysia; 1929 | A. Thompson; Robideau et al. 2011 | HQ643238 | HQ665194 | – | – | HQ708301 | – | |
| P. humicolab | CBS 200.81 | ||||||||||
| ATCC 52179 | – | Soil under Citrus sp. | Taiwan; – | P.J. Ann & W.H. Ko; Robideau et al. 2011 | HQ643243 | HQ665148 | AY564069 | EU080172 | HQ708305 | AY564011 | |
| P. ilicisb | WPC P3939 | ||||||||||
| ATCC 56615 | – | Ilex sp. | British Columbia, Canada; 1988 | H. Ho; Robideau et al. 2011 | HQ261583 | EU079864 | – | – | HQ261330 | – | |
| P. infestansb | CBS 366.51 | – | Solanum tuberosum | The Netherlands; – | –; Robideau et al. 2011 | HQ643247 | HQ665217 | – | – | HQ708309 | – |
| P. kernoviaeb | WPC P10681 | ||||||||||
| ICMP 14761 | – | Annona cherimola | New Zealand; 2002 | C.F. Hill; Robideau et al. 2011 | HQ261603 | EU079650 | – | – | HQ261350 | – | |
| P. litchiib | CBS 100.81 | – | Litchi chinensis | Taiwan; – | C.W. Kao; Voglmayr 2003 | AY198308 | AF235949 | – | – | HQ708323 | – |
| P. megakaryab | WPC P8516 | – | Theobroma cacao | Sao Tome and Principe; – | –; Robideau et al. 2011 | HQ261609 | EU079974 | – | – | HQ261356 | – |
| P. niederhauseriib | WPC P10616 | – | Hedera helix | North Carolina, USA; 2001 | G. Abad; Robideau et al. 2011 | HQ261702 | EU080233 | – | – | HQ261449 | – |
| P. polonicab | WPC P15005 | – | Soil under Alnus glutinosa | Poland; – | T. Oszako; Robideau et al. 2011 | HQ261646 | EU080261 | – | – | HQ261393 | – |
| P. quercinab | WPC P10334 | ||||||||||
| CBS 782.95 | – | Soil and root of decaying | Germany; 1995 | T. Jung; Robideau et al. 2011 | HQ261659 | EU080494 | – | – | HQ261406 | – | |
| Quercus robur | |||||||||||
| P. rubib | CBS 967.95 | ||||||||||
| ATCC 90442 | |||||||||||
| IMI 355974 | – | Rubus idaeus | Scotland; 1985 | J.M. Duncan & D.M. Kennedy; Robideau et al. 2011 | HQ643340 | HQ665306 | KU899234 | KU899391 | HQ708388 | KU899476 | |
| Phytopythium borealeb | CBS 551.88 | – | Soil under Brassica caulorapa | China; – | Y. Yang-nian; Robideau et al. 2011 | HQ643372 | HQ665261 | – | – | HQ708419 | – |
| Ph. helicoidesb | CBS 286.31 | – | Phaseolus vulgaris | USA; – | C. Drechsler; Robideau et al. 2011 | HQ643383 | HQ665186 | – | – | HQ708430 | – |
| Ph. oedochilumb | CBS 292.37 | – | – | USA; – | C. Drechsler; Robideau et al. 2011 | HQ643392 | HQ665191 | – | – | HQ708439 | – |
| Ph. ostracodesb | CBS 768.73 | – | Clay soil | Spain (Ibiza); 1972 | A.J. van der Plaats-Niterink; Robideau et al. 2011 | HQ643395 | HQ665295 | – | – | HQ708442 | – |
| Pythium attrantheridiumb | – | DAOM 230383 | Daucus carota | Canada; – | N. Allain-Boulé; Robideau et al. 2011 | HQ643477 | HQ665308 | – | – | HQ708524 | – |
| Py. caudatumbh | CBS 584.85 | ||||||||||
| ATCC 58383 | – | Xiphinema rivesi | Pennsylvania, USA; 1984 | B.A. Jaffee; Robideau et al. 2011 | HQ643136 | HQ665277 | – | – | HQ708209 | – | |
| Py. insidiosumb | CBS 574.85 | ||||||||||
| ATCC 58643 | – | Horse | Costa Rica; – | –; Robideau et al. 2011 | HQ643570 | HQ665273 | – | – | HQ708614 | – | |
| Py. oligandrumb | CBS 382.34 | – | Matthiola sp. | UK; – | n.a.; Robideau et al. 2011 | HQ643715 | HQ665223 | – | – | HQ708759 | – |
| Py. rostratumb | CBS 533.74 | DAOM 229266 | Soil | The Netherlands; 1971 | A.J. van der Plaats-Niterink; Robideau et al. 2011 | HQ643767 | HQ665252 | – | – | HQ708808 | – |
| Py. ultimum var. ultimumb | CBS 122650 | – | Soil | France; 2012 | T. Rintoul; Robideau et al. 2011 | HQ643864 | HQ665103 | – | – | HQ708905 | – |
| Py. vanterpooliib | CBS 295.37 | – | Triticum aestivum | UK; 1936 | T.C. Vanterpool; Robideau et al. 2011 | HQ643952 | HQ665193 | – | – | HQ708993 | – |
| Salisapilia tartareab | CBS 208.95 | IFO 32606 | Submerged decaying leaf of | Florida, USA; 1991 | S.Y. Newell; Robideau et al. 2011 | HQ643135 | HQ232464 | – | – | HQ708208 | – |
| Spartina alterniflora | |||||||||||
– = not available; authentic strains, ex-types, isotypes, neotypes and paratypes are printed in bolditalics-type.
a Abbreviations of isolates and culture collections: ATCC = American Type Culture Collection, Manassas, USA; CBS = CBS collection at the Westerdijk Fungal Biodiversity Institute (previously Centraalbureau voor Schimmelcultures), Utrecht, Netherlands; DAOM = Canadian National Mycological Herbarium, Agriculture and Agri-Food Canada, Ottawa, Canada; DAR = New South Wales Plant Pathology Herbarium, Orange Agricultural Institute, Orange, Australia; ICMP = International Collection of Micro-organisms from Plants, Auckland, New Zealand; IFO = Institute for Fermentation, Osaka, Japan; WPC = World Phytophthora Collection, University of California Riverside, USA; other isolate names and numbers are as given by the collectors and on GenBank, respectively.
b Isolates used in the phylogenetic studies.
c Isolates used in the morphological studies.
d Isolates used in the temperature-growth studies.
e Submitted to GenBank as Phytophthora sp. REB326-69.
f Submitted to GenBank as Phytophthora sp. PR12-475.
g Submitted to GenBank as Phytophthora sp. PR13-109.
h Submitted to GenBank as Lagenidium caudatum.
DNA isolation, amplification and sequencing
For all Nothophytophthora isolates obtained in this study mycelial DNA was extracted from pure cultures grown in peabroth medium (Erwin & Ribeiro 1996). Pea-broth cultures were kept for 7–10 d at 25 °C without shaking. Mycelium was harvested by filtration through filter paper, washed with sterile deionized water, freeze-dried and ground to a fine powder in liquid nitrogen. Total DNA was extracted using the E.Z.N.A.® Fungal DNA Mini Kit (OMEGA Bio-tek, Norcross, GA) following the manufacturer’s instructions and checked for quality and quantity by spectrophotometry. DNA was stored at –20 °C until further use to amplify and sequence four nuclear and two mitochondrial loci (Table 1). The internal transcribed spacer (ITS1-5.8S-ITS2) region (ITS) and the 5’ terminal domain of the large subunit (LSU) of the nuclear ribosomal RNA gene (nrDNA) were amplified separately using the primer-pairs ITS1/ITS4 (White et al. 1990) and LR0R/LR6-O (Moncalvo et al. 1995, Riethmüller et al. 2002), respectively, using the PCR reaction mixture and cycling conditions described by Nagy et al. (2003) with an annealing temperature of 57 °C (ITS) or 53 °C (LSU) for 30 s. Partial heat shock protein 90 (HSP90) gene was amplified with the primers HSP90F1int and HSP90R1 as described previously (Blair et al. 2008). Segments of the β-tubulin (Btub) and the mitochondrial genes cytochrome c oxidase subunit 1 (cox1) and NADH dehydrogenase subunit 1 (NADH1) were amplified with primers TUBUF2 and TUBUR1, FM80RC (the reverse complement of FM80) and FM85, and NADHF1 and NADHR1, respectively, using the PCR reaction mixture and cycling conditions as described earlier (Martin & Tooley 2003, Kroon et al. 2004). Products of Thermo Fisher Scientific Inc. (Waltham, MA, USA) and Bio-Rad C1000™ or Applied Biosystems® 2720 Thermal Cyclers were used for the PCR reactions. Amplicons were purified and sequenced in both directions using the primers of the PCR reactions by LGC Genomics GmbH (Berlin, Germany). Electrophoregrams were quality checked and forward and reverse reads were compiled using Pregap4 v. 1.5 and Gap v. 4.10 of the Staden software package (Staden et al. 2000). Clearly visible pronounced double peaks were considered as heterozygous positions and labelled according to the IUPAC coding system. All sequences derived in this study were deposited in GenBank and accession numbers are given in Table 1.
Phylogenetic analysis
The sequences obtained in this work were complemented with sequences deposited in GenBank. Four datasets were established to analyse different phylogenetic questions. The sequences of the loci used in the analyses were aligned using the online version of MAFFT v. 7 (Katoh & Standley 2013) by the E-INS-I strategy (ITS) or the auto option (all other loci). When indel positions of ITS sequences were used to increase robustness of phylogenetic analyses (Nagy et al. 2012), the program GapCoder was used (Young & Healy 2003).
To study the (i) phylogenetic position of the potentially new genus among other oomycete genera, a 3-locus dataset (ITS-LSU-cox1) of representative species from all genera of the Peronosporales together with the representatives of all species from the potentially new genus were analysed with Salisapilia tartarea (CBS 208.95), Salisapiliaceae, Peronosporales, Halophytophthora epistomium (CBS 590.85), Peronosporales, and Aphanomyces euteiches (CBS 156.73), Leptolegniaceae, Saprolegniales, as outgroups (dataset: 48 isolates and 3 020 characters). To analyse the (ii) intrageneric phylogeny of the potential new genus a 6-partition dataset (6 loci: ITS-LSU-Btub-HSP90-cox1-NADH1 complemented with the indel motifs of the ITS region) was analysed with Phytophthora boehmeriae (CBS 291.29), P. humicola (CBS 200.81) and P. rubi (CBS 967.95) as outgroup taxa (dataset: 42 isolates and 5 366 characters). A GenBank blast search revealed ITS sequences of three isolates from Ireland and New Zealand which possibly represent congeneric taxa. To analyse their relation to the six new taxa, a (iii) full ITS dataset (complemented with the indel motifs) of all isolates from the six new taxa together with three GenBank entries (dataset: 51 isolates and 1 244 characters) and (iv) a partial ITS dataset (complemented with the indel motifs) of all isolates from the six new taxa together with those three and one partial ITS sequence originating from an environmental sample (MOTU 33 from Català et al. 2015) (dataset: 51 isolates and 1 phylotype; 504 characters) were used. In the ITS datasets P. boehmeriae (CBS 291.29), P. captiosa (P10719), P. kernoviae (P10681) and P. polonica (P15005) were used as outgroup taxa.
A Maximum likelihood (ML) and a Bayesian (BI) analysis were carried out with all datasets except the partial ITS dataset with which only an ML analysis was run (data not shown). Bayesian analyses were performed with MrBayes 3.1.2 (Huelsenbeck & Ronquist 2001, Ronquist & Huelsenbeck 2003) into partitions with GTR+G model for nucleotide partitions and a two-parameter Markov (Mk2 Lewis) model for the indel partitions. Four Markov chains were run for 10 M generations, sampling every 1 000 steps, and with a burn in at 4 000 trees. ML analyses were carried out with the raxmlGUI v. 1.3 (Silvestro & Michalak 2012) implementation of the RAxML (Stamatakis 2014). A GTR+G nucleotide substitution model was used for the nucleotide partitions and indel data were treated as binary data. There were 10 runs of the ML and bootstrap (‘thorough bootstrap’) analyses with 1 000 replicates used to test the support of the branches. Phylogenetic trees were visualized in MEGA6 (Tamura et al. 2013) and edited in figure editor programs. Datasets presented and trees deriving from Maximum likelihood and Bayesian analyses are available from TreeBASE (20801; http://purl.org/phylo/treebase/phylows/study/TB2:S20801).
Morphology of asexual and sexual structures
Morphological features of sporangia, oogonia, oospores, antheridia, chlamydospores, hyphal swellings and aggregations of the six new species (Table 1, 12) were compared with each other.
Table 12.
Morphological characters and dimensions (μm), cardinal temperatures (°C) and temperature-growth relations (mm/d) on V8-juice agara of Nothophytophthora species. Most discriminating characters are highlighted in bold. Percentages in brackets are ranges of isolate means.
| N. amphigynosa | N. caduca | N. chlamydospora | N. valdiviana | N. intricata | N. vietnamensis | |
|---|---|---|---|---|---|---|
| No. of isolates | 8b | 14b | 5b | 5b | 6b | 8b |
| Sporangia | 82 % ovoid, 12 % ellipsoid, 5 % obpyriform (limoniform, mouse-shaped) | 83 % ovoid, 7 % ellipsoid, 4 % limoniform (obpyriform, pyriform, mouse-shaped) | 44 % ovoid, 27.5 % ellipsoid, 22.5 % limoniform (obpyriform, pyriform, mouse-shaped) | 50.5 % ovoid, 40.5 % limoniform, 6 % ellipsoid, (obpyriform, pyriform, mouse-shaped) | 71 % ovoid, 15 % obpyriform, 7 % limoniform, 5 % ellipsoid (pyriform, mouse-shaped) | 91 % ovoid, 6 % ellipsoid, 3 % limoniform |
| l × b mean | 47.0 ± 5.6 × 26.4 ± 1.8 | 37.9 ± 4.6 × 25.7 ± 3.0 | 37.6 ± 4.9 × 22.1 ± 2.5 | 42.7 ± 4.6 × 28.0 ± 3.5 | 38.5 ± 2.8 × 24.8 ± 1.5 | 36.4 ± 12.7 × 29.3 ± 8.1 |
| range of isolate means | 41.5–52.0 × 25.4–27.3 | 34.7–43.1 × 23.3–28.2 | 35.6–38.9 × 20.4–23.2 | 40.4–44.7 × 25.6–29.5 | 37.6–40.5 × 23.4–26.3 | 34.1–37.9 × 24.1–25.8 |
| total range | 33.6–60.6 × 21.3–32.4 | 24.1–54.4 × 18.1–35.9 | 27.4–57.2 × 17.0–30.8 | 30.2–55.7 × 18.6–47.5 | 27.8–49.2 × 18.6–30.2 | 28.4–42.1 × 20.6–28.1 |
| l/b ratio | 1.78 ± 0.17 | 1.48 ± 0.15 | 1.71 ± 0.17 | 1.53 ± 0.14 | 1.55 ± 0.18 | 1.47 ± 0.08 |
| caducity | – | 32.1 % (10–53 %) | 25.2 % (11–41 %) | 6.8 % (4–10 %) | – | 15.8 % (4–36 %) |
| pedicel-like basal plug | 2.9 ± 0.6 | 2.6 ± 0.7 | 2.8 ± 1.6 | 2.4 ± 0.5 | 2.9 ± 0.7 | 2.7 ± 0.7 |
| internal proliferation | – | nested and extended | – | nested and extended | – | – |
| exitpores | 8.9 ± 1.4 | 10.4 ± 2.2 | 8.2 ± 1.7 | 9.4 ± 1.8 | 9.0 ± 1.6 | 7.6 ± 1.5 |
| sympodia | infrequent, lax | frequent, lax | frequent, lax or dense | frequent, lax or dense | infrequent, lax | frequent, lax or dense |
| zoospore cysts | 9.0 ± 1.1 | 7.4 ± 0.6 | 8.6 ± 0.8 | 8.6 ± 1.1 | 8.1 ± 1.1 | 8.4 ± 0.7 |
| sporangiospore swellings | 11.1 ± 2.8; rare | 10.2 ± 2.0; rare | 15.2 ± 6.3; rare | 14.0 ± 2.7; rare | 9.8 ± 1.5; rare | n/a; rare |
| Breeding system | homothallic | self-sterile | self-sterile | self-sterile | homothallic | homothallic |
| Oogonia | ||||||
| mean diam | 25.3 ± 1.7 | – | – | – | 30.1 ± 3.9 | 23.9 ± 3.0 |
| range of isolate means | 24.3–25.5 | – | – | – | 28.1–31.8 | 22.3–27.3 |
| total range | 18.4–29.7 | – | – | – | 16.7–41.8 | 18.6–33.0 |
| tapering base | 2.9 % (0–7.5 %) | – | – | – | 7.5 % (0–30 %) | 75.4 % (42–95 %) |
| thin stalks | 58.3 % (10–100 %) | – | – | – | 29.4 % (2.5–45 %) | 3.1 % (0–12.5 %) |
| curved base | – | – | – | – | 1.3 % (0–5 %) | 24.4 % (7.5–32.5 %) |
| elongated | 12.5 % (5–20 %) | – | – | – | 5.6 % (0–17.5 %) | 70.6 % (60–85 %) |
| Oospores | – | – | – | |||
| plerotic oospores | 99.2 % | – | – | – | 96.9 % (92.5–100 %) | 96.9 % (87.5–100 %) |
| mean diam | 23.4 ± 1.7 | – | – | – | 28.3 ± 3.5 | 22.5 ± 2.4 |
| Total range | 17.2–28.0 | – | – | – | 15.7–38.4 | 17.6–29.5 |
| wall diam | 1.7 ± 0.3 | – | – | – | 2.1 ± 0.4 | 1.8 ± 0.3 |
| oospore wall index | 0.38 ± 0.05 | – | – | – | 0.38 ± 0.06 | 0.42 ± 0.05 |
|
| ||||||
| Abortion rate | 4.2 % (1–25 %) | – | – | – | 10.8 % (1–18 %) | 1.0 % (0–4 %) |
| Antheridia | 87.2 % amphigynous | – | – | – | 100 % paragynous | 100 % paragynous |
| size | 8.5 ± 1.8 × 6.5 ± 0.9 | – | – | – | 10.0 ± 1.9 × 6.9 ± 1.2 | 7.2 ± 1.2 × 4.6 ± 0.9 |
| intricate stalks | 28.8 % (22.5–35 %) | – | – | – | 63.3 % (50–72.5 %) | 46.7 % (42.5–52.5 %) |
| Chlamydospores | – | – | 98.1 % globose, 1.9 % pyriform; radiating, forming clusters 43.7 ± 7.0 | – | – | – |
| Hyphal swellings | – | – | globose, (pyriform, limoniform) 29.2 ± 6.1 | – | – | – |
| Lethal temperature | 28 | 28 or 30 | 26 | 30 | 28 | 29 |
| Maximum temperature | 27 | 26 or 28 | 25 | 28 | 27 | 27 |
| Optimum temperature | 20 | 20 or 25 | 20 | 25 | 25 | 25 |
| Growth rate at 20 °C | 3.1 ± 0.05 | 3.1 ± 0.21 | 3.2 ± 0.05 | 2.9 ± 0.05 | 2.2 ± 0.06 | 2.5 ± 0.04 |
| Growth rate at 25 °C | 3.0 ± 0.06 | 3.6 ± 0.08 | 0.5 ± 0 | 3.1 ± 0.1 | 2.5 ± 0.07 | 2.9 ± 0.05 |
a Oogonia and oospores were studied and measured on carrot agar.
b Numbers of isolates included in the growth tests: N. amphigynosa = 4; N. caduca = 10; N. chlamydospora = 4; N. valdiviana = 4; N. intricata = 5; N. vietnamensis = 8.
– = character not observed.
Formation of sporangia was induced by submersing two 12–15 mm square discs cut from the growing edge of a 3–7-d-old V8A colony in a 90 mm diam Petri dish in non-sterile soil extract (50 g of filtered oak forest soil in 1 000 mL of distilled water, filtered after 24 h) (Jung et al. 1996). The Petri dishes were incubated at 20 °C in natural light and the soil extract was changed after c. 6 h (Jung et al. 2017b). Shape, type of apex, caducity and special features of sporangia and the formation of hyphal swellings and aggregations were recorded after 24–48 h. For each isolate 40 sporangia were measured at ×400 using a compound microscope (Zeiss Imager.Z2), a digital camera (Zeiss Axiocam ICc3) and a biometric software (Zeiss AxioVision).
The formation of chlamydospores and hyphal swellings was examined on V8A after 21–30 d growth at 20 °C in the dark. If present, for each isolate each 40 chlamydospores and hyphal swellings chosen at random were measured under a compound microscope at ×400.
The formation of gametangia (oogonia and antheridia) and their characteristic features were examined after 21–30 d growth at 20 °C in the dark on CA which for oogonia production proved to be superior to V8A in a preliminary study. For each isolate each 40 oogonia, oospores and antheridia chosen at random were measured under a compound microscope at ×400. The oospore wall index was calculated according to Dick (1990). Self-sterile isolates were paired with isolates from the same and from other self-sterile Nothophytophthora species according to Jung et al. (2017b). In addition, isolates from all self-sterile and homothallic Nothophytophthora species were paired with A1 and A2 tester strains of P. cinnamomi using a modified membrane method (Ko et al. 1978, Gallegly & Hong 2008) with nitrocellulose instead of polycarbonate membranes (pore size 0.22 μm; Millipore, Merck, Germany) to test whether they are able to stimulate oogonia production in P. cinnamomi and, hence, share the A1/A2 compatibility system of Phytophthora.
Colony morphology, growth rates and cardinal temperatures
Colony growth patterns of all six Nothophytophthora species were described from 10-d-old cultures grown at 20 °C in the dark in 90 mm plates on CA, V8A, malt-extract agar (MEA; Oxoid Ltd., UK) and potato dextrose agar (PDA; Oxoid Ltd., UK) according to Jung & Burgess (2009), Jung et al. (2017b) and Erwin & Ribeiro (1996).
For temperature-growth relationships, representative isolates of the six Nothophytophthora species (Table 1) were subcultured onto 90 mm V8A plates and incubated for 24 h at 20 °C to stimulate onset of growth (Jung et al. 1999). Then three replicate plates per isolate were transferred to 5, 10, 15, 20, 25, 26, 27, 28, 29 and 30 °C. Radial growth was recorded after 6 d, along two lines intersecting the centre of the inoculum at right angles and the mean growth rates (mm/d) were calculated. To determine the lethal temperature, plates showing no growth at 26, 27, 28, 29 or 30 °C were re-incubated at 20 °C.
RESULTS
Phylogenetic analysis
Compared to the ML analyses the BI analyses provided with all three datasets more support for terminal clades and with the 3-loci dataset also for the deeper branches. Since the topology of all trees resulting from BI and ML analyses was similar the Bayesian trees are presented here with both Bayesian Posterior Probability values and Maximum Likelihood bootstrap values included (Fig. 1, 2, 3, TreeBASE: 20801). When the phylogenetic position of the new genus Nothophytophthora among other oomycete genera was studied with the help of the 3-loci dataset (ITS-LSU-cox1), both BI and ML analyses resulted in a fully supported distinct clade of the isolates of the new genus which formed a monophyletic group with the genus Phytophthora. The clade of the two genera was supported by a 0.98 PP in BI analysis (Fig. 1) and 61 % bootstrap value in ML analysis (not shown). The phylogeny of the other oomycete genera included in the analyses was in accordance with results from previous studies (Hulvey et al. 2010, Marano et al. 2014, Martin et al. 2014, De Cock et al. 2015). The downy mildews, represented by Peronospora rumicis and Hyaloperonospora sisymbrii-sophiae, resided within the paraphyletic genus Phytophthora. The genus Halophytophthora proved to be polyphyletic with Halophytophthora s.str., represented by H. avicenniae, H. batemanensis and H. polymorphica, clustering in a basal position to the monophyletic Phytophthora-Nothophytophthora clade and H. exoprolifera clustering basal to the previous three genera. Halophytophthora operculata resided in a basal position to the genus Phytopythium whereas H. epistomium clearly belongs to an unknown genus outside of the Peronosporaceae and Pythiaceae (Fig. 1). Phytopythium constituted the basal genus within the Peronosporaceae sensu Dick (2001) and Hulvey et al. (2010) which also comprised Halophytophthora, Phytophthora inclusive the downy mildew genera, and Nothophytophthora.
Fig. 1.
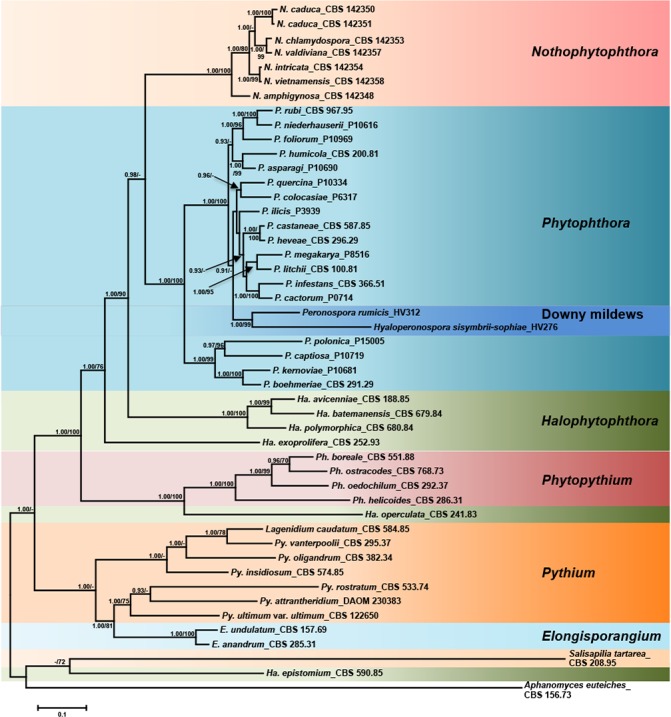
Fifty percent majority rule consensus phylogram derived from Bayesian phylogenetic analysis of three-loci (LSU, ITS, cox1) dataset of Nothophytophthora gen. nov. and representative species from other genera of the Peronosporales. Bayesian posterior probabilities and Maximum Likelihood bootstrap values (in %) are indicated, but not shown below 0.9 and 70 %, respectively. Salisapilia tartarea, Halophytophthora epistomium and Aphanomyces euteiches were used as outgroup taxa. Scale bar indicates 0.1 expected changes per site per branch.
Fig. 2.
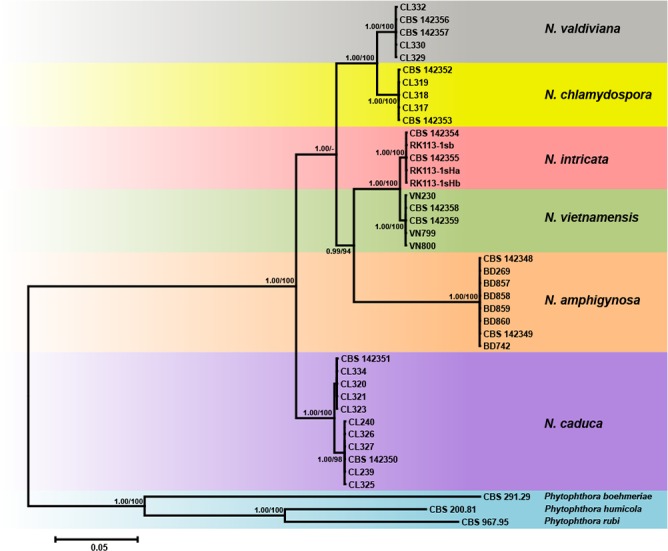
Fifty percent majority rule consensus phylogram derived from Bayesian phylogenetic analysis of six-loci (LSU, ITS, Btub, HSP90, cox1, NADH1) dataset of Nothophytophthora gen. nov. to examine intrageneric variability and phylogenetic structure. Bayesian posterior probabilities and Maximum Likelihood bootstrap values (in %) are indicated, but not shown below 0.9 and 70 %, respectively. Phytophthora boehmeriae, P. humicola and P. rubi were used as outgroup taxa. Scale bar indicates 0.05 expected changes per site per branch.
Fig. 3.
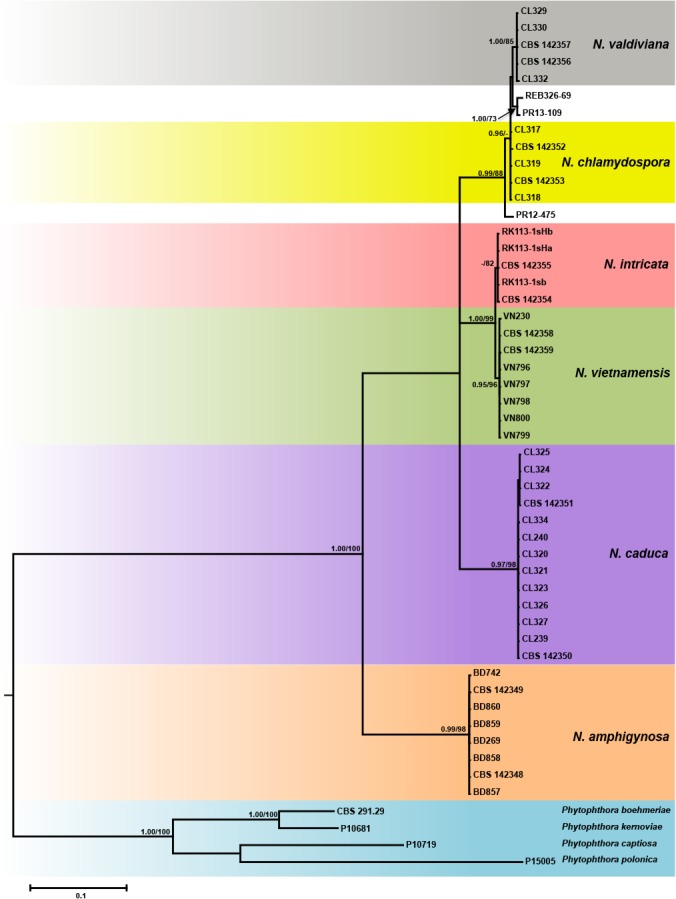
Fifty percent majority rule consensus phylogram derived from Bayesian inference analysis of a full ITS dataset (complemented with the indel motifs) of the six new Nothophytophthora species and three GenBank entries from Ireland and New Zealand. Bayesian posterior probabilities and Maximum Likelihood bootstrap values (in %) are indicated, but not shown below 0.9 and 70 %, respectively. Phytophthora boehmeriae, P. captiosa, P. kernoviae and P. polonica were used as outgroup taxa. Scale bar indicates 0.1 expected changes per site per branch.
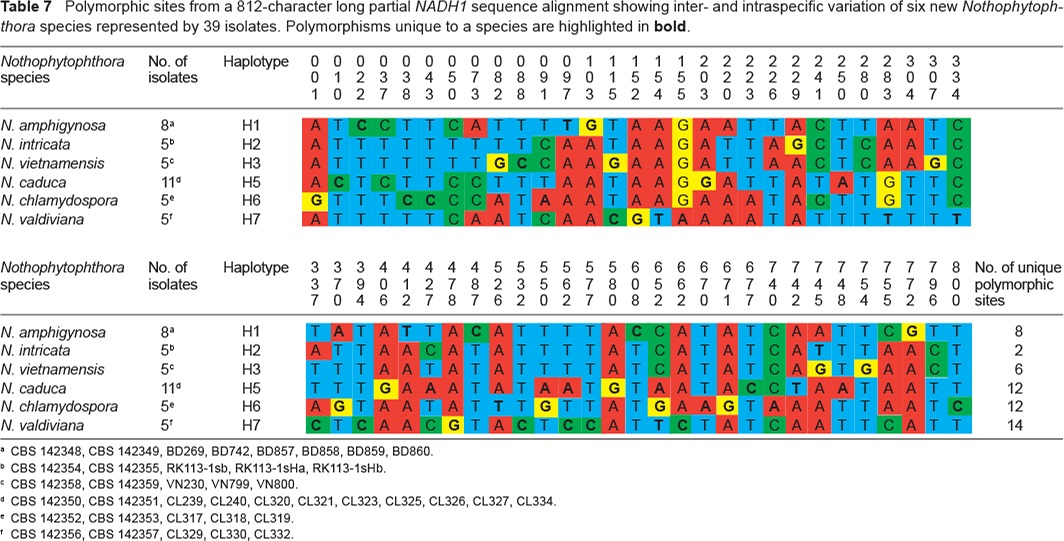
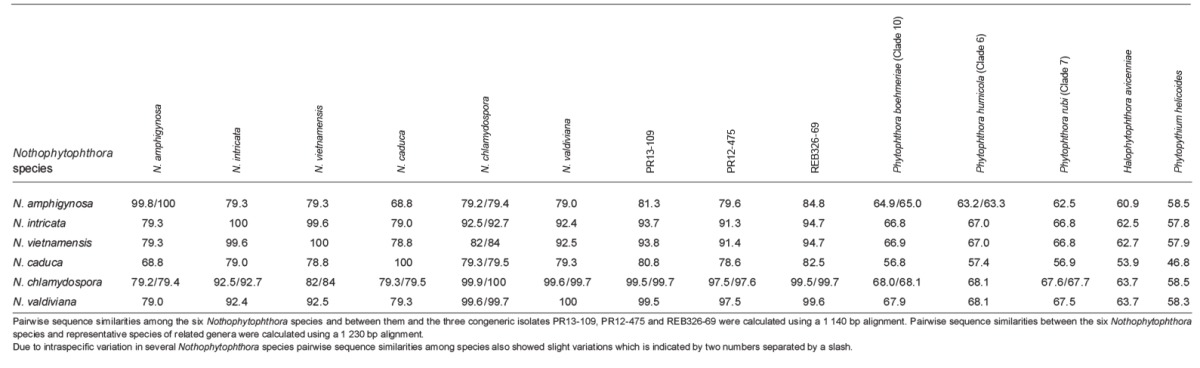
When the intrageneric phylogeny of Nothophytophthora was analysed with the 6-partition dataset (ITS-LSU-Btub-HSP90-cox1-NADH1), the isolates formed six fully supported distinct clades (Fig. 2). In both BI and ML analyses the two populations of N. caduca from two different forest streams formed two separate clusters within the N. caduca clade which resided in a basal position to the well-supported clade formed by the other five Nothophytophthora species (Fig. 2). Within that clade, N. vietnamensis with N. intricata and N. valdiviana with N. chlamydospora clustered in sister position to each other with both subclades having high support in both analyses. However, the relative position of these two clades and the position of the fifth lineage, N. amphigynosa, have been fully resolved only in the BI analysis (Fig. 2). Across a 4 136 character alignment of the five coding genes, LSU, Btub, HSP90, cox1 and NADH1, N. amphigynosa, N. intricata, N. vietnamensis, N. caduca, N. chlamydospora and N. valdiviana had 31, 7, 9, 53, 29 and 31 unique polymorphisms, respectively, and differed from each other at 19–116 positions corresponding to sequence similarities of 97.2–99.5 % (Table 8, 9). The six Nothophytophthora species differed from Phytophthora spp. (P. boehmeriae, P. humicola and P. rubi), Halophytophthora avicenniae and Phytopythium helicoides at 328–379, 370–382 and 472–491 positions corresponding to sequence similarities of 90.8–92.1 %, 90.8–91.0 % and 88.1–88.6 % (Table 8, 9). Due to the presence of heterozygous positions N. amphigynosa and N. chlamydospora had four and two LSU haplotypes, respectively (Table 3). Also, the LSU sequence of all isolates of N. vietnamensis contained one heterozygous position (Table 3). Heterozygous sites were also present in the ITS sequences of N. amphigynosa, N. chlamydospora and N. vietnamensis (Table 2) and in the HSP90 sequences of N. caduca and N. chlamydospora (Table 4). The Btub sequence of all isolates of N. valdiviana contained nine heterozygous positions (Table 5). No heterozygous positions were found in the mitochondrial cox1 and NADH1 sequences of any Nothophytophthora isolate (Table 6, 7).
Table 8.
Pairwise numbers of different positions along a 4 136-character long multigene alignment (LSU, Btub, HSP90, cox1, NADH1) among the six Nothophytophthora species and between the Nothophytophthora species and representative species of the related genera Phytophthora, Halophytophthora and Phytopythium.
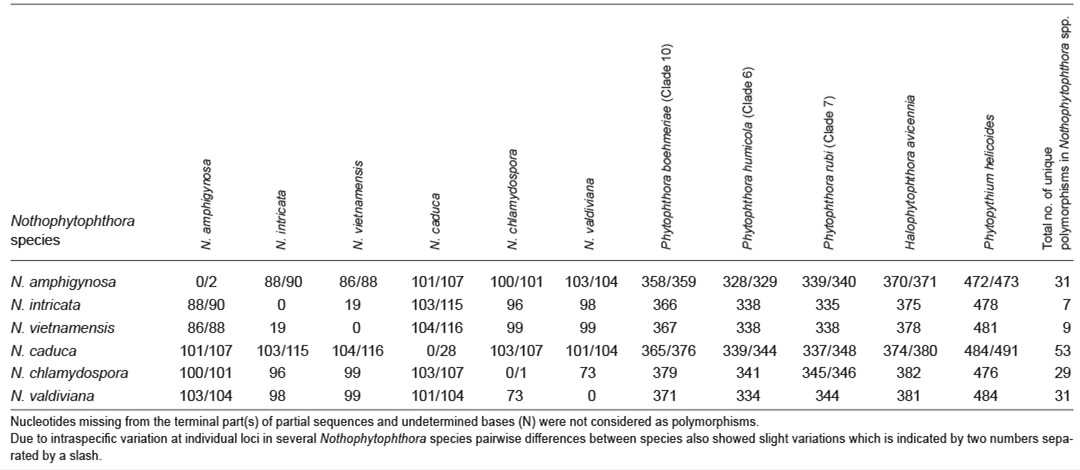
Table 9.
Pairwise sequence similarities (%) along a 4 136-character long multigene alignment (LSU, Btub, HSP90, cox1, NADH1) among the six Nothophytophthora species and between the Nothophytophthora species and representative species of the related genera Phytophthora, Halophytophthora and Phytopythium.
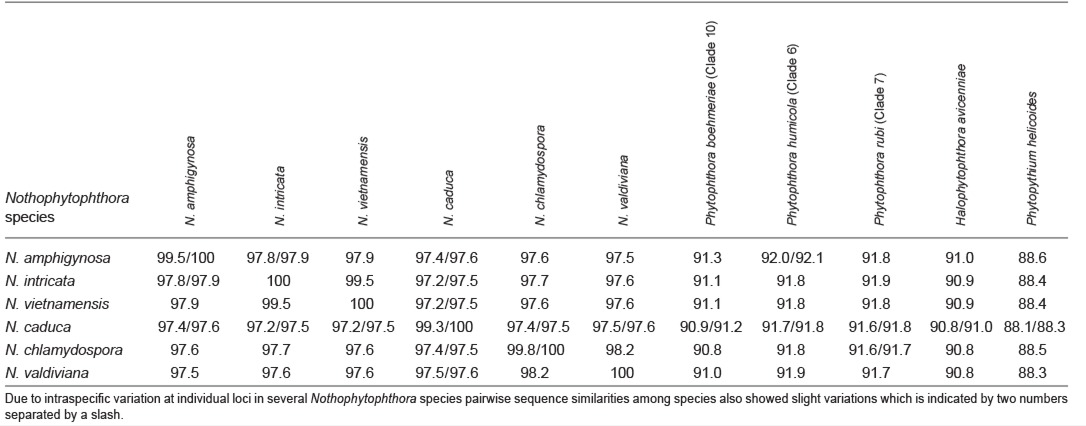
Table 3.
Polymorphic sites from a 986-character long partial LSU sequence alignment showing inter- and intraspecific variation of 6 new Nothophytophthora species represented by 45 isolates. Polymorphisms unique to a species are highlighted in bold.
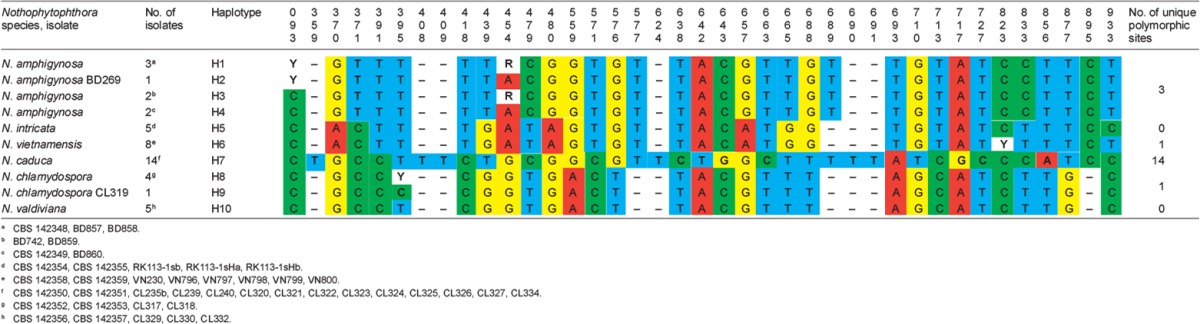
Table 2.
Polymorphic sites from a 1 140-character long ITS rDNA sequence alignment showing inter- and intraspecific variation of the six new Nothophytophthora species represented by 45 isolates. Polymorphisms unique to a species are highlighted in bold.
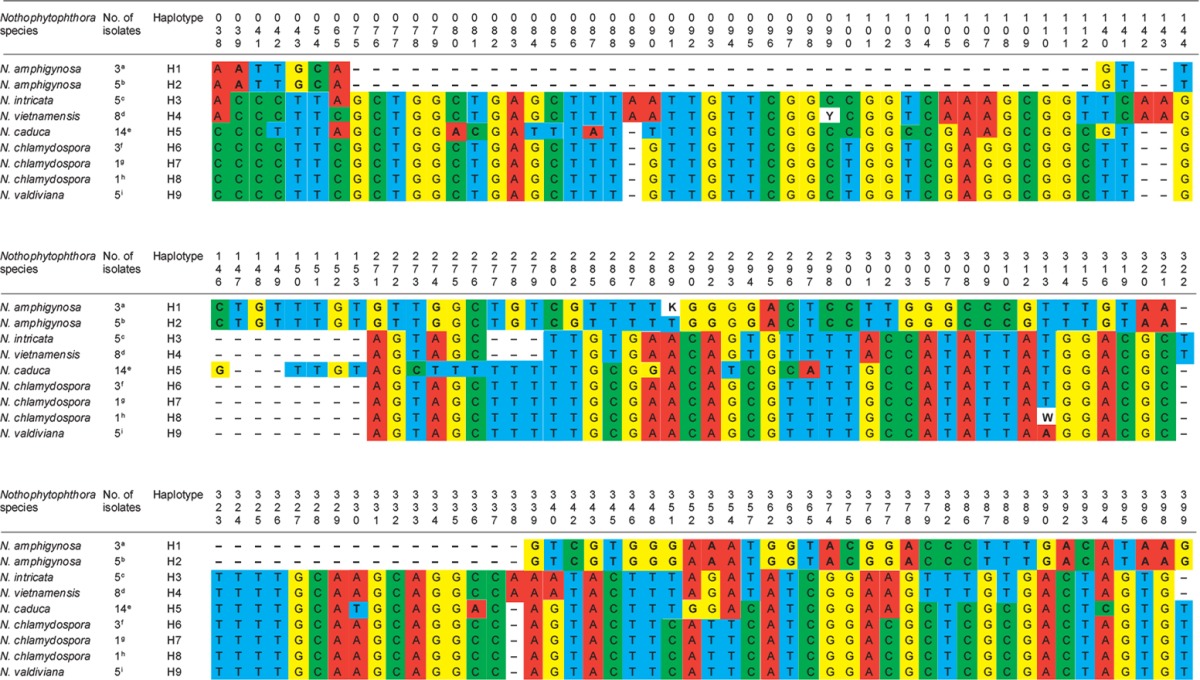
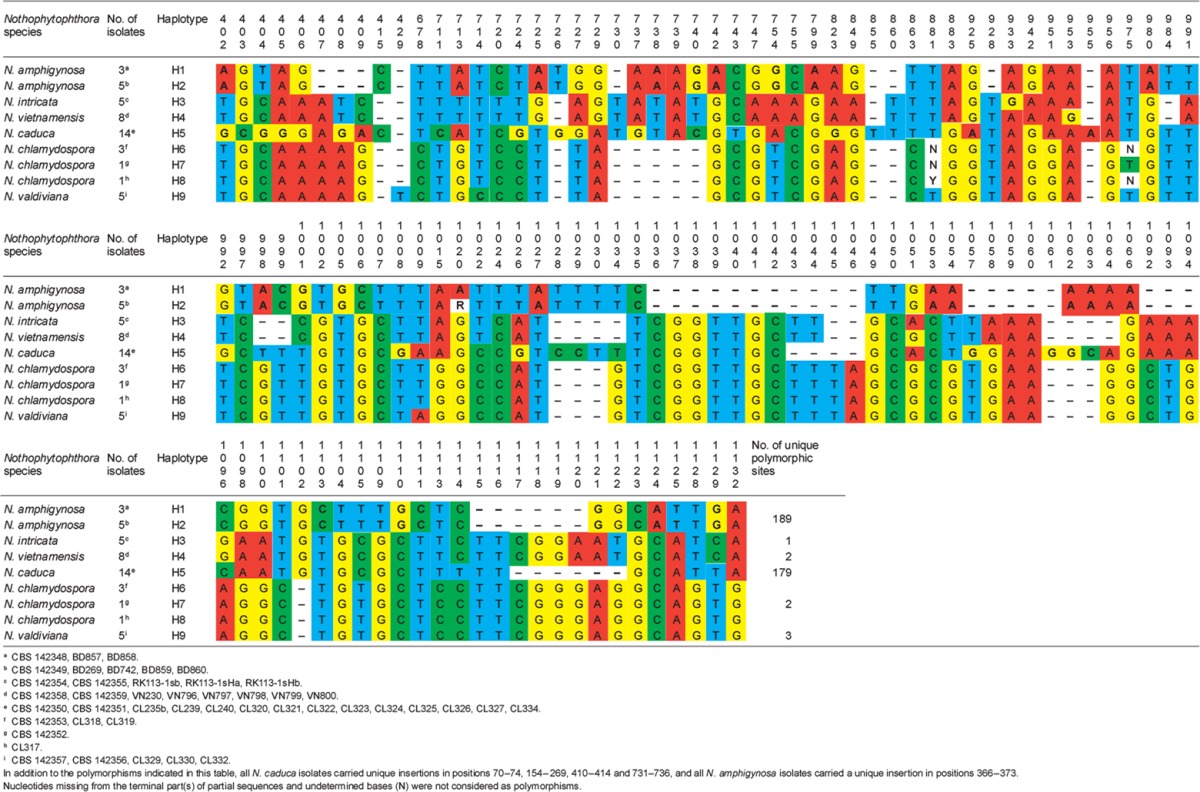
Table 4.
Polymorphic sites from a 833-character long partial HSP90 sequence alignment showing inter- and intraspecific variation of six new Nothophytophthora species represented by 39 isolates. Polymorphisms unique to a species are highlighted in bold.
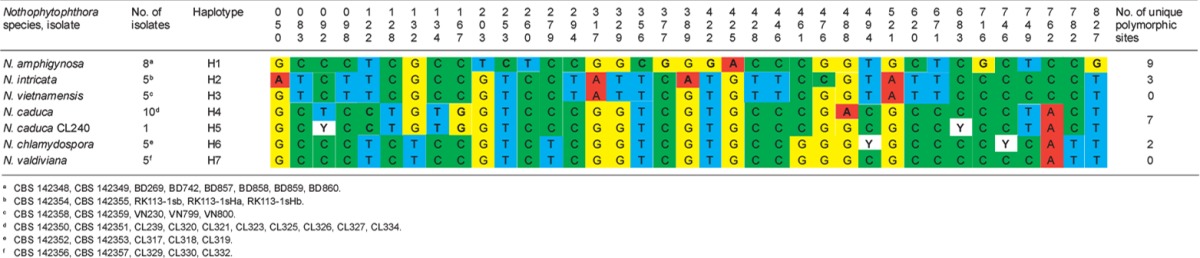
Table 5.
Polymorphic sites from a 897-character long partial ß-tubulin sequence alignment showing inter- and intraspecific variation of six new Nothophytophthora species represented by 39 isolates. Polymorphisms unique to a species are highlighted in bold.
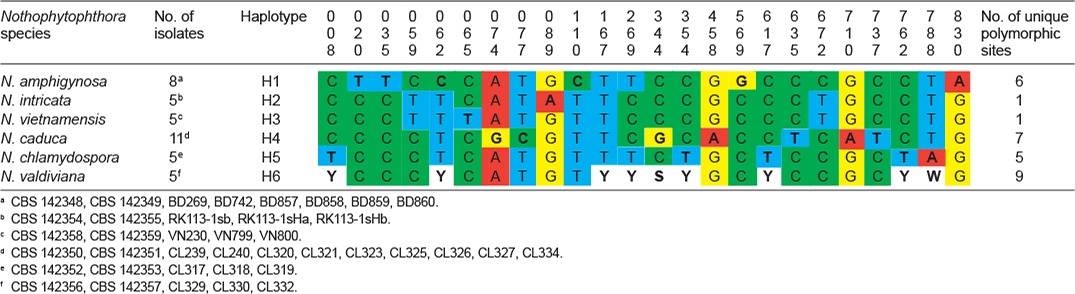
Table 6.
Polymorphic sites from a 643-character long partial cox1 sequence alignment showing inter- and intraspecific variation of six new Nothophytophthora species represented by 39 isolates. Polymorphisms unique to a species are highlighted in bold.
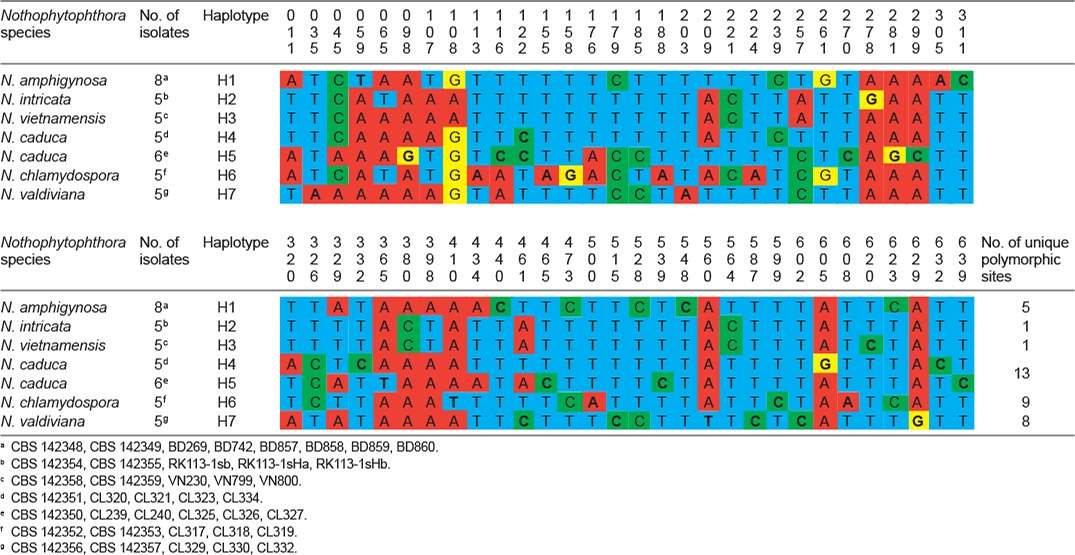
Table 7.
Polymorphic sites from a 812-character long partial NADH1 sequence alignment showing inter- and intraspecific variation of six new Nothophytophthora species represented by 39 isolates. Polymorphisms unique to a species are highlighted in bold.
When the ITS sequences of the six new Nothophytophthora species together with three (partial) ITS sequences from GenBank were analysed the phylogeny gained was less supported in both BI and ML analyses (Fig. 3). Nothophytophthora amphigynosa clustered in a basal position to the other five Nothophytophthora species of which the relative positions could not be fully resolved (Fig. 3). The ITS sequences of the three congeneric isolates from streams in Ireland and New Zealand grouped into the clade formed by N. valdiviana and N. chlamydospora with the Irish isolate PR12-475 being basal to this clade. Isolates PR13-109 from Ireland and REB326-69 from New Zealand showed only 4–6 characters differences to N. chlamydospora and N. valdiviana (Table 10) but their phylogenetic position was vague and they could not unambiguously be assigned to any of the two new species (Fig. 3). In a separate analysis of a shorter ITS sequence alignment (data not shown), a similar situation was found. The partial ITS sequence representing the ‘Uncultured Phytophthora clone R2 MOTU33’ from a metagenomic stream survey in Spain (Català et al. 2015) grouped within the clade formed by the sister species N. vietnamensis and N. intricata. Unfortunately, the short ITS sequence, even complemented with the indels, could not resolve the clade of these two new species (data not shown) and the species assignation of this environmental sequence was ambiguous (NB: the sequence of ‘MOTU 33’ was not included in the analyses presented in Català et al. 2015).
Table 10.
Pairwise numbers of different positions along ITS rDNA sequence alignments among the six Nothophytophthora species and between the Nothophytophthora species, the congeneric three isolates PR13-109, PR12-475 and REB326-69 and representative species of the related genera Phytophthora, Halophytophthora and Phytopythium.
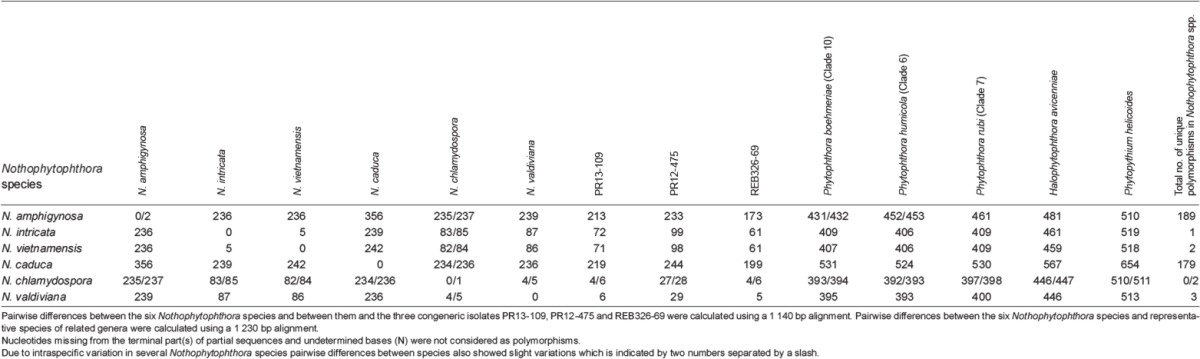
In the ITS region with its non-coding parts, both intrageneric variability and differences between Nothophytophthora and related genera were considerably higher than in the coding genes. In the 1 140 bp ITS alignment used for the intrageneric comparison the sequences of the six Nothophytophthora species contained in total 417 polymorphic sites (36.6 %; Table 2) and showed pairwise differences at 5–356 positions, equivalent to sequence similarities of 68.8–99.7 % (Table 10, 11). The large differences of N. amphigynosa and N. caduca to other Nothophytophthora spp. were caused by the high numbers of 189 and 179 unique polymorphisms, respectively, which mainly comprised indels (Table 2). Including the three congeneric isolates from Ireland and New Zealand increased the number of polymorphic sites to 427 (data not shown). In the 1 230 characters ITS alignment used for the intergeneric comparison the six Nothophytophthora species differed from Phytophthora spp. (P. boehmeriae, P. humicola and P. rubi), H. avicenniae and Ph. helicoides at 392–531, 446–567 and 510–654 positions corresponding to sequence similarities of 56.8–68.1 %, 53.9–63.7 % and 46.8–58.5 % (Table 10, 11).
Table 11.
Pairwise sequence similarities along ITS rDNA alignments among the six Nothophytophthora species and between the Nothophytophthora species, the three congeneric isolates PR13-109, PR12-475 and REB326-69 and representative species of the related genera Phytophthora, Halophytophthora and Phytopythium.
TAXONOMY
Nothophytophthora T. Jung, Scanu, Bakonyi & M. Horta Jung, gen. nov. — MycoBank MB820530
Etymology. Name refers to the morphological and ecological similarity to Phytophthora (Nothus Lat = false).
Type species. Nothophytophthora amphigynosa.
Sporangia mostly ovoid, limoniform, ellipsoid or obpyriform, and usually non-papillate. Sporangial proliferation is in all known species external, leading in some species to dense sympodia, and in some species also internal in both a nested and extended way. In all known species, a conspicuous opaque plug separates most sporangia from the sporangiophores. In some species, varying proportions of the sporangia are caducous, breaking off at the base of this plug which is synonymous with the pedicel of airborne Phytophthora species. As in Phytophthora, the sporangial cytoplasm differentiates inside the sporangia into biflagellate zoospores which are released without discharge tube. Chlamydospores are formed in some species and are absent in others. Some species are homothallic, forming smooth-walled oogonia, containing thick-walled oospores with a large ooplast, and amphigynous and/or paragynous antheridia. Several species are sterile both in single culture and when mated with isolates from the same or from other self-sterile Nothophytophthora species, and also when mated with A1 and A2 tester strains of Phytophthora cinnamomi. Nothophytophthora is morphologically similar to Phytophthora and phylogenetically constitutes a monophyletic sister genus of Phytophthora.
Notes — Morphological and physiological characters and morphometric data of the six new Nothophytophthora species are given in the comprehensive Table 12.
Nothophytophthora amphigynosa T. Jung, Scanu, Bakonyi & M. Horta Jung, sp. nov. — MycoBank MB820532; Fig. 4
Fig. 4.
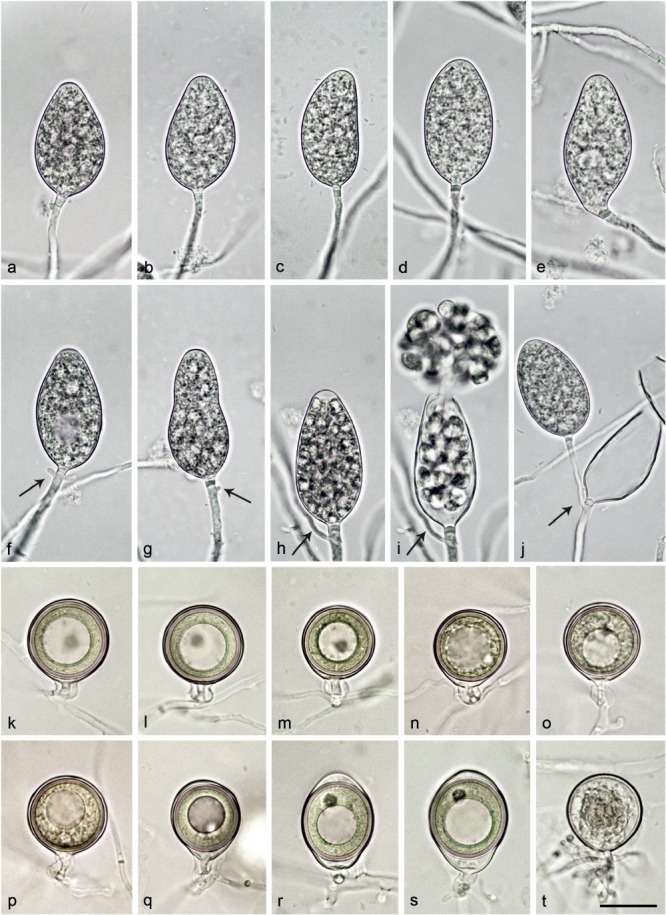
Morphological structures of Nothophytophthora amphigynosa. — a–j. Non-papillate sporangia with a conspicuous basal plug formed on V8 agar flooded with soil extract. a–e. Sporangia borne terminally on unbranched sporangiophores; a–c. ovoid; c. with a slightly curved apex; d. ellipsoid; e. limoniform with slightly lateral attachment of the sporangiophore; f–j. sporangia with external proliferation immediately below sporangial base (arrows); f. ovoid with vacuole; g. elongated obpyriform with slightly lateral attachment of the sporangiophore; h. elongated-ovoid with already differentiated zoospores; i. same sporangium as in h releasing zoospores; j. lax sympodium of two ovoid sporangia. — k–p. Smooth-walled mature oogonia with non-tapering bases and short, thin stalks, containing plerotic, medium thick-walled oospores with each one large ooplast and one nucleus, formed in single culture in V8A; k–o. globose to subglobose with amphigynous antheridia; p. globose with paragynous antheridium behind oogonial stalk; q. slightly elongated with tapering base and amphigynous antheridium; r–s. elongated-ellipsoid with tapering bases, slightly elongated almost plerotic oospores and amphigynous antheridia; t. smooth-walled oogonium aborted before forming an oospore. — Scale bar = 25 μm, applies to a–t.
Etymology. Name refers to the predominantly amphigynous antheridia.
Typus. Portugal, Sintra, isolated from a stream in a temperate Atlantic forest, T. Jung, 13 Mar. 2015 (CBS H-23007 holotype, dried culture on CA, Herbarium Westerdijk Fungal Biodiversity Institute, CBS 142348 = BD268, ex-type culture). ITS and cox1 sequences GenBank KY788382 and KY788473, respectively.
Sporangia, hyphal swellings and chlamydospores (Fig. 4a–j) — Sporangia were not observed in solid agar but were produced abundantly in non-sterile soil extract. Sporangia of N. amphigynosa were typically borne terminally on unbranched sporangiophores (Fig. 4a–e) or less frequently in lax sympodia of 1–3 sporangia (Fig. 4f–j), and some were formed intercalary (0.3 %). Small subglobose to limoniform hyphal swellings (11.1 ± 2.8 μm) were sometimes observed on sporangiophores. Sporangia were non-caducous and non-papillate (Fig. 4a–j). In almost all mature sporangia (98.5 %) a conspicuous opaque plug was formed inside the sporangiophore close to the sporangial base which averaged 2.9 ± 0.6 μm (Fig. 4a–j). Sporangia were mostly ovoid to elongated-ovoid (over all isolates 81.5 %; Fig. 4a–c, f, h–j), ellipsoid (11.6 %; Fig. 4d, j), obpyriform (5.1 %; Fig. 4g) or infrequently limoniform (0.9 %; Fig. 4e), mouse- or club-shaped (0.9 %). Sporangia with special features such as slightly lateral attachment of the sporangiophore (over all isolates 14.1 %; Fig. 4e, g), slightly curved apex (3.1 %; Fig. 4c) or the presence of a vacuole (5.9 %; Fig. 4f) were common in all isolates. Sporangial proliferation was exclusively external (28.8 % of sporangia; Fig. 4f–j). Sporangial dimensions of eight isolates of N. amphigynosa averaged 47.0 ± 5.6 × 26.4 ± 1.8 μm (overall range 33.6–60.6 × 21.3–32.4 μm) with a range of isolate means of 41.5–52.0 × 25.4–27.3 μm and a length/breadth ratio of 1.78 ± 0.17 (range of isolate means 1.62–1.91) (Table 12). Zoospores of N. amphigynosa were differentiated inside the sporangia and discharged through an exit pore 5.2–16.3 μm wide (av. 8.9 ± 1.4 μm) (Fig. 4h–j). They were limoniform to reniform whilst motile, becoming spherical (av. diam = 9.0 ± 1.1 μm) on encystment. Direct germination of sporangia was not observed. In solid agar, hyphal swellings or chlamydospores were not observed.
Oogonia, oospores and antheridia (Fig. 4k–t) — Gametangia were produced in single culture by all isolates of N. amphigynosa in CA within 10–14 d. Gametangia formation was usually starting at and was sometimes restricted to the areas of the colonies close to the walls of the Petri dishes. Oogonia were borne terminally, had smooth walls, short thin stalks and were globose to slightly subglobose with non-tapering bases (on av. 87.5 %; Fig. 4k–p, t) or less frequently elongated pyriform to ellipsoid (12.5 %) sometimes with tapering bases (2.9 %) (Fig. 4q–s). Mean diameter of oogonia was 25.3 ± 1.7 μm (overall range 18.4–29.7 μm and range of isolate means 24.3–25.5 μm). They were almost exclusively plerotic (99.2 %). Oospores of N. amphigynosa were usually globose (Fig. 4k–q) but could be slightly elongated in elongated oogonia (Fig. 4r–s). Oospores contained large ooplasts (Fig. 4k–s) and had a diameter of 23.4 ± 1.7 μm (overall range 17.2–28.0 μm), a wall diam of 1.7 ± 0.3 μm (range 1.0–2.5 μm) and an oospore wall index of 0.38 ± 0.05 (Table 12). Oospore abortion was low (4.2 % after 4 wk; Fig. 4t). The antheridia often had twisted intricate stalks (28.8 %) and were club-shaped to subglobose, mostly amphigynous (87.2 %; Fig. 4k–o, q–s) or less frequently paragynous (12.8 %; Fig. 4p) and averaged 8.5 ± 1.8 × 6.5 ± 0.9 μm. In the nitrocellulose membrane test all isolates tested stimulated abundant oogonia production in the A2 tester strain of P. cinnamomi.
Colony morphology, growth rates and cardinal temperatures (Fig. 10, 11) — Colonies on V8A, CA and MEA were largely submerged with limited aerial mycelium around the inoculum plug. They had a chrysanthemum pattern on V8A and CA and were uniform on MEA. On PDA colonies were dense felty with a rosaceous pattern (Fig. 10). Temperature-growth relations are shown in Fig. 11. All four isolates included in the growth test had similar growth rates and cardinal temperatures. The maximum growth temperature was 27 °C. The isolates did not resume growth when plates incubated for 5 d at 28 °C were transferred to 20 °C. The average radial growth rates at the optimum temperature of 20 °C and at 25 °C were 3.1 ± 0.05 and 3.0 ± 0.06 mm/d, respectively (Fig. 11).
Fig. 10.

Colony morphology of Nothophytophthora amphigynosa, N. caduca, N. chlamydospora, N. valdiviana, N. intricata and N. vietnamensis (from top to bottom) after 10 d growth at 20 °C on V8 agar, carrot agar, potato-dextrose agar and malt extract agar (from left to right).
Fig. 11.
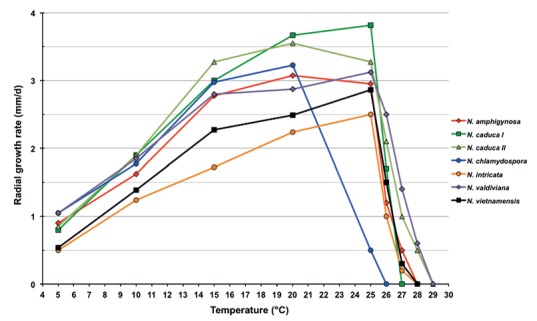
Mean radial growth rates of Nothophytophthora amphigynosa (4 isolates), N. caduca (6 isolates from population N. caduca I; 4 isolates from population N. caduca II), N. chlamydospora (4 isolates), N. intricata (5 isolates), N. valdiviana (4 isolates) and N. vietnamensis (8 isolates) on V8 agar at different temperatures.
Additional specimens. Portugal, Sintra, isolated from a stream in a temperate Atlantic forest, T. Jung, 13 Mar. 2015; CBS 142349 = BD741; BD269; BD742; BD857; BD858; BD859; BD860.
Nothophytophthora caduca T. Jung, Scanu, Bakonyi, A. Durán & M. Horta Jung, sp. nov. — MycoBank MB820534; Fig. 5
Fig. 5.
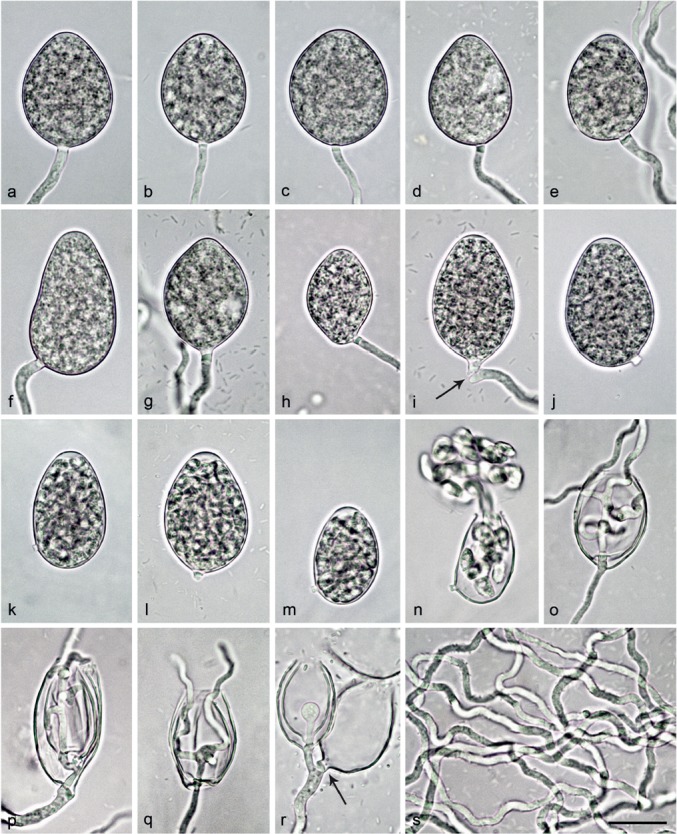
Morphological structures of Nothophytophthora caduca formed on V8 agar flooded with soil extract. — a–j. Mature non-papillate sporangia; a. ovoid without basal plug; b–n. with conspicuous basal plug; b. ovoid; c–e. ovoid with undulating sporangiophores; f. obpyriform with undulating sporangiophore; g. limoniform with undulating sporangiophore; h. limoniform with laterally attached sporangiophore; i. ovoid, just being shed (arrow); j–n. caducous ovoid sporangia with short pedicel-like basal plug; k–m. with differentiated zoospores and swollen semipapillate apex; n. same sporangium as in m releasing zoospores; o–q. empty sporangia with internal nested and extended proliferation and multiple branching and undulating growth of hyphae inside the sporangium; r. small sympodium of two sporangia resulting from external proliferation; one sporangium showing nested proliferation and the other one breaking off from the sporangiophore (arrow); s. undulating hyphae. — Scale bar = 25 μm in a–r and 40 μm in s.
Etymology. Name refers to the caducity of the sporangia (caduca Lat = caducous, shedding).
Typus. Chile, isolated from a stream in a temperate Valdivian rainforest, T. Jung, 25 Nov. 2014 (CBS H-23011 holotype, dried culture on CA, Herbarium Westerdijk Fungal Biodiversity Institute, CBS 142350 = CL328, ex-type culture). ITS and cox1 sequences GenBank KY788401 and KY788489, respectively.
Sporangia, hyphal swellings and chlamydospores (Fig. 5a–r) — Sporangia of N. caduca were not observed in solid agar but were produced abundantly in non-sterile soil extract. Sporangia were borne terminally on unbranched sporangiophores or less frequently in lax sympodia of 1–3 sporangia (Fig. 5r). A conspicuous, pedicel-like opaque plug (2.6 ± 0.7 μm) formed inside the sporangiophore close to the base of most sporangia (over all isolates 87.0 %; Fig. 5b–n, r). In all isolates, sporangia were partially caducous (10–53 %, on av. 32.1 %; Fig. 5i–n) breaking off at the base of the basal plug. Sporangial shapes ranged from broadly ovoid, ovoid or elongated ovoid (83.4 %; Fig. 5a–e, i–j, l, o–r) to ellipsoid (7.4 %; Fig. 5k, m–n), limoniform (4.1 %; Fig. 5g–h), mouse-shaped (3.0 %), obpyriform (15.3 %; Fig. 5f), subglobose (0.7 %) or pyriform (0.7 %). Sporangia with laterally attached sporangiophores (44.6 %; Fig. 5e–f, h, j–k, m–n) and undulating sporangiophores (74.1 %; Fig. 5d–g, i, r–s) were commonly observed. Sporangial proliferation was external (Fig. 5r) and internal in both a nested and extended way (Fig. 5o–r) often with the sporangiophore showing multiple branching and undulating growth inside the empty sporangium (Fig. 5o–q). Sporangial dimensions of 14 isolates of N. caduca averaged 37.9 ± 4.6 × 25.7 ± 3.0 μm (overall range 24.1–54.4 × 18.1–35.9 μm) with a range of isolate means of 34.7–43.1 × 23.3–28.2 μm. The length/breadth ratio averaged 1.48 ± 0.15 with a range of isolate means of 1.38–1.66 (Table 12).Germination was indirect with zoospores (Fig. 5n) discharged through an exit pore 4.3–16.9 μm wide (av. 10.4 ± 2.2 μm; Fig. 5n–r). They were limoniform to reniform whilst motile, becoming spherical (av. diam = 7.4 ± 0.6 μm) on encystment. Subglobose to limoniform swellings were infrequently formed on sporangiophores. Chlamydospores were not observed.
Oogonia, oospores and antheridia — All 14 isolates of N. caduca were self-sterile and did not form gametangia when paired against each other or with isolates of N. chlamydospora, N. valdiviana and with A1 and A2 tester strains of P. cinnamomi. Since in the nitrocellulose membrane test all isolates tested stimulated abundant oogonia production in the A2 tester strain of P. cinnamomi, their breeding system was considered as silent A1 mating type.
Colony morphology, growth rates and cardinal temperatures (Fig. 10, 11) — All isolates of N. caduca formed similar colonies on the same agar medium. Colonies on V8A, CA and PDA had a rosaceous to chrysanthemum pattern, largely submerged with limited felty aerial mycelium around the inoculum on V8A and CA and more woolly on PDA. On MEA irregular to dendroid, dense-felty colonies were formed (Fig. 10). The temperature-growth relations on V8A are shown in Fig. 11. The two populations from different streams had slightly different optimum and maximum temperatures for growth of 25 and 26 °C in one population and 20 and 28 °C in the other population (Fig. 11). Lethal temperatures were 28 and 30 °C, respectively. All isolates showed slow growth with average radial growth rates of 3.1 ± 0.2 mm/d at 20 °C and 3.6 ± 0.08 mm/d at 25 °C (Fig. 11).
Additional specimens. Chile, isolated from streams in a temperate Valdivian rainforest, T. Jung, 25 Nov. 2014; CBS 142351 = CL333; CL235b; CL239; CL240; CL320; CL321; CL322; CL323; CL324; CL325; CL326; CL327; CL334.
Nothophytophthora chlamydospora T. Jung, Scanu, Bakonyi, A. Durán & M. Horta Jung, sp. nov. — MycoBank MB820536; Fig. 6
Fig. 6.
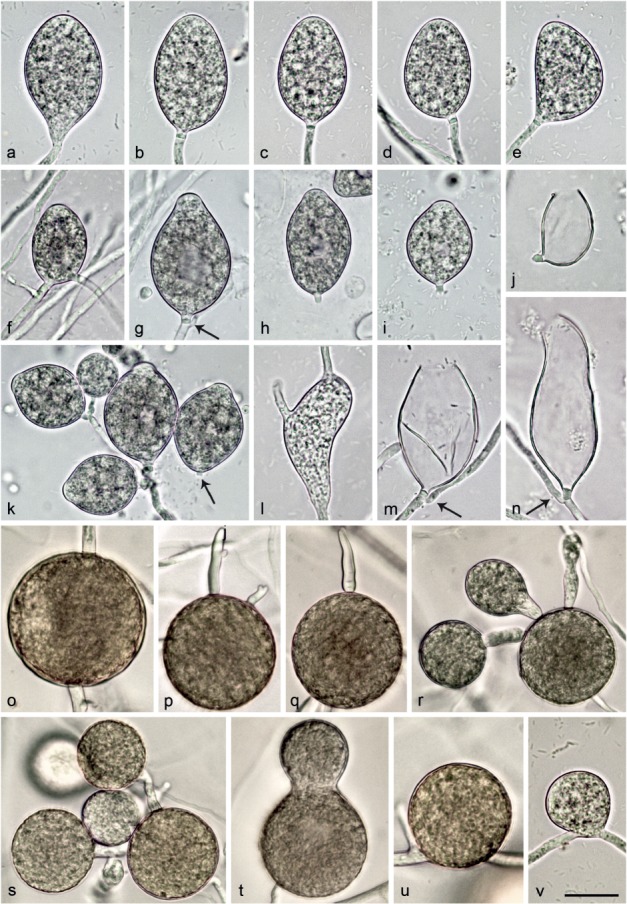
Morphological structures of Nothophytophthora chlamydospora. — a–n. Structures formed on V8 agar flooded with soil extract; a–i. mature non-papillate sporangia; a–e. borne terminally on unbranched sporangiophores; a. ellipsoid with tapering base; b–e. with conspicuous basal plugs; b. ellipsoid; c. ovoid; d. ovoid with slight lateral attachment of sporangiophore; e. mouse-shaped with laterally attached sporangiophore; f. ovoid, intercalary inserted; g. ovoid with vacuole, basal plug and beginning external proliferation (arrow); h–i. caducous with short pedicel-like basal plugs and small vacuoles; h. ellipsoid; i. limoniform; j. ovoid caducous sporangium with short pedicel-like basal plug, after release of zoospores; k. dense sympodium of ovoid to limoniform sporangia with shallow semipapillate apices; one sporangium caducous with short pedicel-like basal plug (arrow); l. sporangium which failed to form a basal septum and continued to grow at the apex, functionally becoming a hyphal swelling; m–n. empty sporangia after release of zoospores, with conspicuous basal plugs and external proliferation close to the base; m. ovoid; n. elongated-obpyriform with curved apex; o–v. structures formed in solid V8 agar; o–u. chlamydospores; o. globose, intercalary inserted; p–q. globose, terminally inserted with hyphal outgrowths; r–s. globose with radiating hyphae forming hyphal swellings or secondary chlamydospores; t. ampulliform, terminally inserted; u. globose, laterally sessile; v. intercalary globose hyphal swelling. — Scale bar = 25 μm, applies to a–v.
Etymology. Name refers to the production of chlamydospores by all known isolates.
Typus. Chile, isolated from a stream in a temperate Valdivian rainforest, T. Jung, 25 Nov. 2014 (CBS H-23008 holotype, dried culture on CA, Herbarium Westerdijk Fungal Biodiversity Institute, CBS 142353 = CL316, ex-type culture). ITS and cox1 sequences GenBank KY788405 and KY788493, respectively.
Sporangia, hyphal swellings and chlamydospores (Fig. 6a–v) — Sporangia of N. chlamydospora were not observed on solid agar but were produced abundantly after 24 hr in non-sterile soil extract. Sporangia were borne terminally (Fig. 6a–e, g) or infrequently intercalary (Fig. 6f) on unbranched sporangiophores or in dense sympodia (Fig. 6k). Up to 6–8 sporangia per sympodium were observed although there were usually fewer. Sporangia were non-papillate or sometimes shallow semi-papillate (Fig. 6k) and partially caducous (over all isolates 11–41 %, on av. 25.2 %; Fig. 6h–k) breaking off below a pedicel-like opaque plug formed inside the sporangiophore close to the base of 77.5 % of all sporangia (Fig. 6b–e, g–k, m–n). Sporangial shapes ranged from ovoid or elongated ovoid (44 %; Fig. 6c–d, f–g, j–k, m), ellipsoid (27.5 %; Fig 6a–b, h) and limoniform (22.5 %; Fig. 6i, k) to obpyriform (2.5 %), mouse-shaped (1.5 %; Fig. 6e) or pyriform (1.5 %). Sporangia with special features like lateral attachment of the sporangiophore (14.5 %; Fig. 6d–e, j), curved apex (2.0 %; Fig. 6n), hyphal extensions (1.5 %; Fig. 6l), a vacuole (13.0 %; Fig. 6g–i) or undulating sporangiophores (2.0 %) occurred in all isolates. Sporangia proliferated exclusively externally, usually immediately below the old sporangium (Fig. 6g, k, m–n). Sporangial dimensions of five isolates averaged 37.6 ± 4.9 × 22.1 ± 2.5 μm (overall range 27.4–57.2 × 17.0–30.8 μm and range of isolate means 35.6–38.9 × 20.4–23.2). The length/breadth ratio averaged 1.71 ± 0.17 with a range of isolate means of 1.64–1.75 (Table 12). In all isolates, a few sporangia failed to form a basal septum and continued to grow at the apex, functionally becoming hyphal swellings (Fig. 6l). Zoospores were discharged through an exit pore 4.8–13.1 μm wide (av. 8.2 ± 1.7 μm; Fig. 6j, m–n). They were limoniform to reniform whilst motile, becoming spherical (av. diam = 8.6 ± 0.8 μm) on encystment. Cysts germinated directly. Intercalary or terminal, globose or limoniform, sometimes catenulate hyphal swellings, measuring 29.2 ± 6.1 μm, were formed by all isolates (Fig. 6v). Globose (98.1 %) or less frequently pyriform to irregular (1.9 %) chlamydospores (Fig. 6o–u) were produced intercalary or terminally and measured 43.7 ± 7.0 μm (Table 12). They often had radiating hyphae bearing hyphal swellings or secondary chlamydospores, thus, forming small clusters of chlamydospores and swellings (Fig. 6p–s).
Oogonia, oospores and antheridia — All five isolates of N. chlamydospora were self-sterile and did not form gametangia when paired with each other or with isolates of N. chlamydospora, N. valdiviana and with A1 and A2 tester strains of P. cinnamomi. Since in the nitrocellulose membrane test all isolates stimulated abundant oogonia production in the A2 tester strain of P. cinnamomi, their breeding system was considered as silent A1 mating type.
Colony morphology, growth rates and cardinal temperatures (Fig. 10, 11) — Colonies on V8A had a striate to chrysanthemum pattern and were largely submerged with very limited aerial mycelium. On CA and MEA colonies with limited aerial mycelium were produced, petaloid on CA and uniform to faintly petaloid on MEA. Colonies on PDA were rosaceous with dense-felty to woolly aerial mycelium (Fig. 10). Temperature-growth relations are shown in Fig. 11. All four isolates included in the growth test had similar growth rates and cardinal temperatures. The maximum and lethal growth temperatures were 25 and 26 °C, respectively. The average radial growth rate at the optimum temperature of 20 °C was 3.2 ± 0.05 mm/d (Fig. 11).
Additional specimens. Chile, isolated from a stream in a temperate Valdivian rainforest, T. Jung, 25 Nov. 2014; CBS 142352 = CL195; CL317; CL318; CL319.
Nothophytophthora intricata T. Jung, Scanu, Bakonyi & M. Horta Jung, sp. nov. — MycoBank MB820538; Fig. 7
Fig. 7.
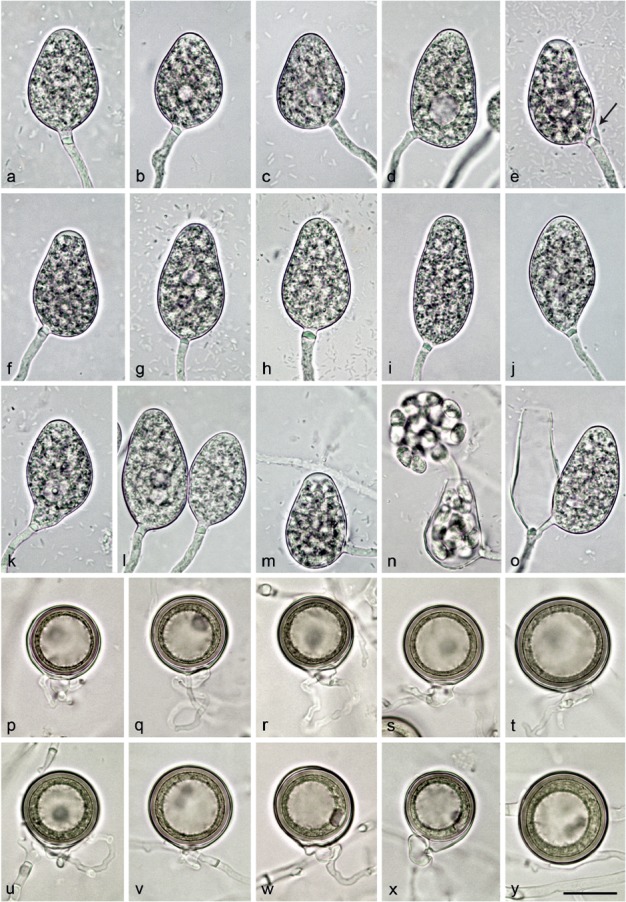
Morphological structures of Nothophytophthora intricata. — a–o. Non-papillate sporangia with conspicuous basal plugs formed on V8 agar flooded with soil extract; a–n. borne terminally on unbranched sporangiophores; a–d. ovoid to elongated-ovoid; b–d. with small vacuoles; c–d. with laterally attached sporangiophores; e–h. obpyriform; e–f. with laterally attached sporangiophores and swollen apices before release of zoospores; i. elongated-obpyriform; j. ellipsoid with tapering slightly curved base; k. pyriform with vacuole and curved base; l. elongated-ovoid, one with vacuoles and laterally attached sporangiophore; m. ovoid with laterally attached sporangiophore and swollen apex, before release of zoospores; n. same sporangium as in m releasing zoospores; o. small sympodium of two sporangia resulting from external proliferation, empty sporangium elongated obpyriform and the other ovoid with laterally attached sporangiophore; p–y. mature, smooth-walled globose oogonia formed in single culture in CA, containing thick-walled plerotic oospores with particularly big ooplasts; p–x. with paragynous antheridia; p–w. with non-tapering bases; p–r. terminally inserted on thin stalks; q–r. with twisting intricate antheridial stalks; u–x. laterally inserted, sessile or on very short stalks; u. with undulating antheridial stalk; w. with twisting intricate antheridial stalk; x. with tapering base; y. intercalary inserted. — Scale bar = 25 μm, applies to a–y.
Etymology. Name refers to the intricate, intertwining antheridial stalks (intricata Lat = intricate or intertwining).
Typus. Germany, Wiesbaden, rhizosphere of a declining mature Aesculus hippocastanum tree in the floodplain of the river Main, T. Jung, 5 Aug. 2011 (CBS H-23009 holotype, dried culture on CA, Herbarium Westerdijk Fungal Biodiversity Institute, CBS 142354 = RK113-1s, ex-type culture). ITS and cox1 sequences GenBank KY788413 and KY788501, respectively.
Sporangia, hyphal swellings and chlamydospores (Fig. 7a–o) — Sporangia were not observed in solid agar but were produced abundantly in non-sterile soil extract. Sporangia of N. intricata were typically borne terminally on unbranched sporangiophores (Fig. 7a–n) or less frequently forming lax sympodia of 1–3 sporangia (5.8 %; Fig. 7e, o). Subglobose to limoniform hyphal swellings (9.8 ± 1.5 μm) were sometimes formed on sporangiophores. Sporangia were non-papillate and non-caducous (Fig. 7a–o). Mature sporangia were usually delimited by a conspicuous opaque plug (91.1 %; 2.9 ± 0.7 μm) formed inside the sporangiophore close to the sporangial base (Fig. 7a–o) which sometimes protruded into the empty sporangium (Fig. 7o). Sporangia with special features such as lateral attachment of the sporangiophore (40.0 %; Fig. 7c–f, m–o), curved base (4.6 %; Fig. 7j–k), curved apex (2.1 %) or the presence of a vacuole (20.0 %; Fig. 7b–e, k, l) and undulating sporangiophores (10.4 %) were common in all isolates. Sporangia were mostly ovoid to elongated-ovoid (70.5 %; Fig. 7a–d, k–o), obpyriform (15.4 %; Fig. 7e–i, o), limoniform (6.3 %), ellipsoid (5.0 %; Fig. 7j) and less frequently pyriform (1.3 %), ampulliform (0.8 %) or mouse-shaped (0.7 %). Sporangial proliferation was exclusively external (Fig. 7e, o). Sporangial dimensions of six isolates of N. intricata averaged 38.5 ± 2.8 × 24.8 ± 1.5 μm (overall range 27.8–49.2 × 18.6–30.2 μm) with a range of isolate means of 37.6–40.5 × 23.4–26.3 μm and a length/breadth ratio of 1.55 ± 0.18 (range of isolate means 1.47–1.65) (Table 12). Zoospores of N. intricata were discharged through an exit pore 4.8–13.8 μm wide (av. 9.0 ± 1.6 μm) (Fig. 7n, o). They were limoniform to reniform whilst motile, becoming spherical (8.1 ± 1.1 μm) on encystment. Direct germination of sporangia was not observed. In solid agar, hyphal swellings or chlamydospores were not formed.
Oogonia, oospores and antheridia (Fig. 7p–y) — Gametangia were readily produced in single culture on CA by all isolates of N. intricata within 10–14 d. Gametangia formation was usually starting at and was sometimes restricted to the edges of the colonies close to the walls of the Petri dishes. Oogonia had smooth walls and were borne terminally on thin, often undulating stalks (Fig. 7p–t) or were sessile (Fig. 7u–x) or less frequently intercalary inserted (Fig. 7y). They were usually globose to slightly subglobose (94.4 %) with mostly non-tapering bases (Fig. 7p–w, y) or less frequently slightly elongated (5.6 %) with tapering bases (Fig. 7x) and almost exclusively plerotic (96.9 %; Fig. 7p–y). Mean diameter of oogonia was 30.1 ± 3.9 μm with an overall range of 16.7–41.8 μm and a range of isolate means of 28.1–31.8 μm (Table 12). Oospores of N. intricata were globose and contained particularly large ooplasts (Fig. 7p–y). Oospore dimensions averaged 28.3 ± 3.5 μm (overall range 15.7–38.4 μm) with a wall diam of 2.1 ± 0.4 μm (range 1.0–3.2 μm) and an oospore wall index of 0.38 ± 0.05 (Table 12). Oospore abortion was low (4.5 % after 4 wk at 20 °C increasing to 10.8 % after 12 mo storage at 8 °C). The antheridia were club-shaped to subglobose and exclusively paragynous (Fig. 7p–y). Antheridial stalks were often intricate and undulating (63.3 %; Fig. 7q–r, u, w). Antheridial dimensions averaged 10.0 ± 1.9 × 6.9 ± 1.2 μm. In the nitrocellulose membrane test all isolates stimulated abundant oogonia production in the A2 tester strain of P. cinnamomi.
Colony morphology, growth rates and cardinal temperatures (Fig. 10, 11) — All isolates of N. intricata formed similar colonies on the same agar medium. Colonies on all media were round with regular margins (Fig. 10). On V8A faintly stellate colonies with limited aerial mycelium were formed. On CA and MEA colonies had stellate patterns and were dense-felty on CA and largely submerged with limited aerial mycelium on MEA. Colonies on PDA were faintly striate with moderate aerial mycelium (Fig. 10).Temperature-growth relations are shown in Fig. 11. All five isolates included in the growth test had similar growth rates and cardinal temperatures. The maximum and lethal temperatures were 27 and 28 °C, respectively. The average radial growth rates at 20 °C and at the optimum temperature of 25 °C were 2.2 ± 0.06 and 2.5 ± 0.07 mm/d, respectively (Fig. 11).
Additional specimens. Germany, Wiesbaden, rhizosphere of declining mature Aesculus hippocastanum trees in the floodplain of the river Main, T. Jung, 5 Aug. 2011; CBS 142355 = RK113-1sH; RK113-1sa; RK113-1sb; RK113-1sHa; RK113-1sHb.
Nothophytophthora valdiviana T. Jung, Scanu, Bakonyi, A. Durán & M. Horta Jung, sp. nov. — MycoBank MB820539; Fig. 8
Fig. 8.
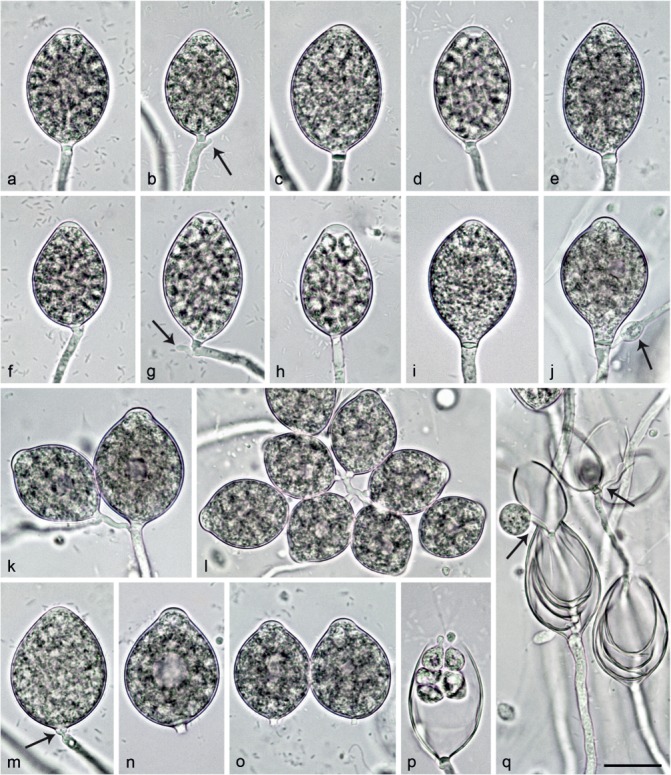
Sporangia of Nothophytophthora valdiviana formed on V8 agar flooded with soil extract. — a–p. Mature non-papillate to shallow semipapillate sporangia with conspicuous basal plugs; a–f. ovoid; a–b, d. with differentiated zoospores to be released soon; b. with beginning external proliferation close to the sporangial base (arrows); g. ellipsoid with beginning external proliferation, just breaking off at base of pedicel-like basal plug; h. obpyriform, just before zoospore release; i. limoniform; j. limoniform with vacuole, external proliferation and swelling on the sporangiophore (arrow); k. small sympodium of two ovoid to limoniform sporangia with vacuoles, resulting from external proliferation; l. dense sympodium of ovoid and mostly limoniform sporangia, some with small vacuoles; m. ovoid sporangium breaking off at base of pedicel-like basal plug (arrow); n–o. ovoid, caducous sporangia with vacuoles and short pedicel-like basal plugs; p. same sporangium as in d releasing zoospores; q. empty sporangia showing internal nested and extended proliferation, and external proliferation (arrows). — Scale bar = 25 μm, applies to a–q.
Etymology. Name refers to the origin of all known isolates in Valdivian rainforests.
Typus. Chile, isolated from a stream in a temperate Valdivian rainforest, T. Jung, 25 Nov. 2014 (CBS H-23010 holotype, dried culture on CA, Herbarium Westerdijk Fungal Biodiversity Institute, CBS 142357 = CL331, ex-type culture). ITS and cox1 sequences GenBank KY788417 and KY788505, respectively.
Sporangia, hyphal swellings and chlamydospores (Fig. 8a–q) — Sporangia of N. valdiviana were not formed on solid agar but were produced abundantly in non-sterile soil extract. Sporangia were non-papillate to shallow semi-papillate (Fig. 8a–o). In all isolates, a low proportion of sporangia were caducous (4–10 %, on av. 6.8 %) breaking off below a pedicel-like opaque plug (2.4 ± 0.5 μm; Fig. 8m–o) which is formed inside the sporangiophore close to the base of 78 % of all sporangia (Fig. 8a–p). Sporangia were ovoid to elongated ovoid (51.0 %; Fig. 8a–f, k–q), limoniform (40.5 %; Fig. 8i–l) and less frequently ellipsoid (6.0 %; Fig. 8g, q) or obpyriform (1.5 %; Fig. 8h). Sporangiophores were sometimes undulating (15.0 %) and infrequently laterally attached to the sporangia (5.5 %). A vacuole was observed in 28.1 % of sporangia (Fig. 8j–l, n, o). Mean sporangial dimensions of five isolates were 42.7 ± 4.6 × 28.0 ± 3.5 μm (overall range 30.2–55.7 × 18.6–47.5 μm) with a range of isolate means of 40.4–44.7 × 25.6–29.5 μm (Table 12). The length/breadth ratio averaged 1.53 ± 0.14 with a range of isolate means of 1.48–1.62. In all isolates, sporangia proliferated internally in both a nested and extended way (Fig. 8q). In addition, sporangiophores often branched externally close to the sporangial base (Fig. 8b, g, j–l, q) forming lax or dense sympodia of up to 8–10 sporangia (Fig. 8k–l). Subglobose to limoniform swellings, averaging 14.0 ± 2.7 μm, were infrequently produced on sporangiophores (Fig. 8j) by all isolates. Zoospores of N. valdiviana were discharged through exit pores 9.4 ± 1.8 μm (5.5–13.0 μm) wide (Fig. 8p–q). They were limoniform to reniform whilst motile (Fig. 8p), becoming spherical (av. diam = 8.6 ± 1.1 μm) on encystment. Cysts germinated directly. Chlamydospores were not observed.
Oogonia, oospores and antheridia — All five isolates of N. valdiviana were self-sterile and did not form gametangia when paired with each other or with isolates of N. caduca, N. chlamydospora and A1 and A2 tester strains of P. cinnamomi. In the nitrocellulose membrane test all isolates stimulated abundant oogonia production in the A2 tester strain of P. cinnamomi. Therefore, their breeding system was considered as silent A1 mating type.
Colony morphology, growth rates and cardinal temperatures (Fig. 10, 11) — All N. valdiviana isolates formed similar colonies on the same agar medium. Colonies on CA and V8A were largely submerged with limited aerial mycelium, with a chrysanthemum pattern on V8A and a stellate to chrysanthemum pattern on CA. On PDA and MEA colonies were appressed with dense felty aerial mycelium and rosaceous patterns, respectively (Fig. 10). The temperature-growth relations on V8A are shown in Fig. 11. All isolates showed slow growth and had similar growth rates at the same temperature. Optimum and maximum growth temperatures were 25 and 28 °C, respectively. Isolates did not resume growth when plates incubated for 5 d at 30 °C were transferred to 20 °C. The average radial growth rates at 20 and 25 °C were 2.9 ± 0.05 mm/d and 3.1 ± 0.1 mm/d, respectively (Fig. 11).
Additional specimens. Chile, isolated from a stream in a temperate Valdivian rainforest, T. Jung, 25 Nov. 2014; CBS 142356 = CL242; CL329; CL330; CL332.
Nothophytophthora vietnamensis T. Jung, Scanu, Bakonyi, P.Q. Thu & M. Horta Jung, sp. nov. — MycoBank MB820541; Fig. 9
Fig. 9.
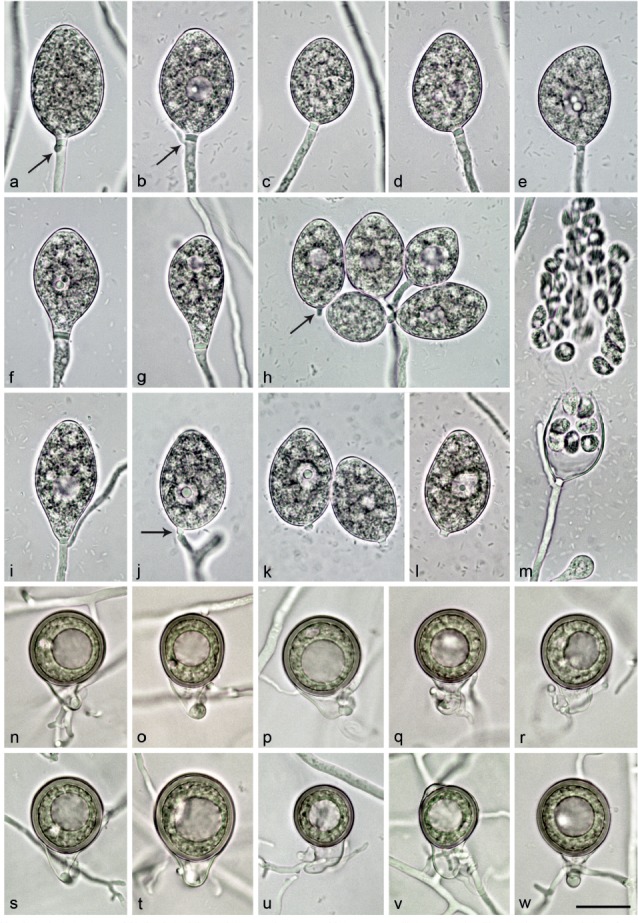
Morphological structures of Nothophytophthora vietnamensis. — a–m. Mature non-papillate sporangia with conspicuous basal plugs formed on V8 agar flooded with soil extract; a–e. ovoid; a–b. with beginning external proliferation close to sporangial base (arrows); b. with vacuole; d. with slightly lateral attachment of sporangiophore; e. with vacuole and curved apex; c–g. borne terminally on unbranched sporangiophores; f. limoniform with vacuole; g. pyriform with vacuole; h. dense sympodium of ellipsoid and ovoid sporangia with vacuoles, one sporangium caducous with pedicel-like basal plug (arrow); i. elongated-pyriform with vacuole and external proliferation; j. ovoid sporangium with vacuole and external proliferation, breaking off at base of pedicel-like basal plug (arrow); k–l. ovoid, caducous sporangia with vacuoles and short pedicel-like basal plugs; m. ovoid sporangium with external proliferation, releasing 40 zoospores; n–w. mature, smooth-walled oogonia formed in single culture in CA, with thick-walled plerotic oospores containing big ooplasts and with paragynous antheridia; n–v. elongated-pyriform with tapering curved bases; o, t. with elongated oospores; n, p, r, v. with undulating antheridial stalks; w. subglobose with short tapering base. — Scale bar = 25 μm, applies to a–w.
Etymology. Name refers to the origin of all known isolates in Vietnam.
Typus. Vietnam, Fansipan, rhizosphere soil of Castanopsis sp. and Acer campbellii, T. Jung, 27 Mar. 2016 (CBS H-23012 holotype, dried culture on CA, Herbarium Westerdijk Fungal Biodiversity Institute, CBS 142358 = VN794, ex-type culture). ITS and cox1 sequences GenBank KY788420 and KY788508, respectively.
Sporangia, hyphal swellings and chlamydospores (Fig. 9a–m) — Sporangia were not observed in solid agar but were produced abundantly in non-sterile soil extract. Sporangia of N. vietnamensis were borne terminally on unbranched sporangiophores (Fig. 9c–g) or in lax or dense sympodia of 2–7 sporangia (Fig. 9h). Small subglobose to limoniform hyphal swellings were only rarely observed on sporangiophores. Mature sporangia were non-papillate and usually delimited by a pedicel-like, conspicuous opaque plug (2.7 ± 0.7 μm) formed inside the sporangiophore close to the sporangial base (over all isolates 93.5 %; Fig. 9a–m). Sporangia were partially caducous (4–36 %, on av. 15.8 %) breaking off just below the basal plug (Fig. 9h, j–l). Sporangial shapes were mostly ovoid to elongated-ovoid (90.5 %; Fig. 9a–e, h, j–m) or infrequently ellipsoid (6.0 %; Fig. 9h), limoniform (3.4 %; Fig. 9f) or pyriform (0.1 %; Fig. 9g, i). Special features such as the presence of a vacuole (55.0 %; Fig. 9b, d–l), slightly lateral attachment of the sporangiophore (14.0 %; Fig. 9c–d, j–l), a curved apex (0.4 %; Fig. 9e) or undulating sporangiophores (3.1 %) were common in all isolates. Sporangial dimensions of eight isolates of N. vietnamensis averaged 36.4 ± 12.7 × 29.3 ± 8.1 μm with an overall range of 28.4–42.1 × 20.6–28.1 μm, a range of isolate means of 34.1–37.9 × 24.1–25.8 μm and a length/breadth ratio of 1.47 ± 0.08 (range of isolate means 1.42–1.52) (Table 12). Sporangial proliferation was exclusively external (20.5 % of sporangia; Fig. 9a–b, h–j, m). Zoospores of N. vietnamensis were discharged through an exit pore 4.1–11.7 μm wide (7.6 ± 1.5 μm) (Fig. 9m). They were limoniform to reniform whilst motile, becoming spherical (8.4 ± 0.7 μm) on encystment. Direct germination of sporangia was not observed. In solid agar, no hyphal swellings or chlamydospores were produced.
Oogonia, oospores and antheridia (Fig. 9n–w) — Gametangia were readily produced in single culture by all isolates of N. vietnamensis on CA within 10–14 d. Gametangia formation was usually starting at and was sometimes restricted to the edges of the colonies close to the walls of the Petri dishes. Oogonia were borne terminally, had smooth walls and due to their mostly long tapering (75.4 %) and curved (24.4 %) bases were mostly elongated pyriform to ellipsoid (70.6 %; Fig. 9n–v) or less frequently globose to slightly subglobose (49.4 %; Fig. 9w). Only 3.1 % of oogonia had particularly thin stalks. Oogonia were small with a mean diameter of 23.9 ± 3.0 μm (overall range 18.6–33.0 μm and range of isolate means 22.3–27.3 μm) (Table 12). They were almost exclusively plerotic (96.9 %). Oospores of N. vietnamensis contained large ooplasts and were usually globose (Fig. 9n, p–s, u, w) or less frequently slightly elongated (10.6 %; Fig. 9o, t, v). Oospores had diameters of 22.5 ± 2.4 μm (overall range 17.6–29.5 μm) with walls averaging 1.8 ± 0.3 μm (range 1.1–2.5 μm) and an oospore wall index of 0.42 ± 0.05 (Table 12). Oospore abortion was low (1.0 % after 4 wk). The antheridia were club-shaped to subglobose, exclusively paragynous (Fig. 9n–w) and small with dimensions averaging 7.2 ± 1.2 × 4.6 ± 0.9 μm. Antheridial stalks were often twisted and undulating (46.7 %; Fig. 9n, p, r, v). In the nitrocellulose membrane test none of the isolates tested stimulated oogonia production in the A1 and A2 tester strains of P. cinnamomi.
Colony morphology, growth rates and cardinal temperatures (Fig. 10, 11) — All isolates produced similar colonies on the same agar medium. Colonies on V8A and MEA were radiate to slightly radiate with limited aerial mycelium around the inoculum plug and a submerged edge on V8A (Fig. 10). On CA, appressed radiate colonies with limited aerial mycelium were formed whereas colonies on PDA had a chrysanthemum pattern with dense-felty aerial mycelium (Fig. 10). Temperature-growth relations are shown in Fig. 11. All eight isolates included in the growth test had similar growth rates and cardinal temperatures. The maximum growth temperature was 27 °C. The isolates did not resume growth when plates incubated for 5 d at 29 °C were transferred to 20 °C. The average radial growth rates at 20 °C and at the optimum temperature of 25 °C were 2.5 ± 0.04 and 2.9 ± 0.05 mm/d, respectively (Fig. 11).
Additional specimens. Vietnam, Fansipan, rhizosphere soil of Castanopsis sp. and Acer campbellii, 27 Mar. 2016, T. Jung; CBS 142359 = VN795; VN230; VN796; VN797; VN798; VN799; VN800.
NOTES
The genera Nothophytophthora and Phytophthora share numerous morphological characters like persistent and caducous sporangia with variable shapes, internal differentiation of zoospores, and external and internal nested and extended sporangial proliferation; smooth-walled oogonia with amphigynous and/or paragynous attachment of the antheridia; chlamydospores and hyphal swellings. Several of these characters are also common to Halophytophthora but general absence of caducity and internal proliferation of sporangia and of amphigynous antheridia, and the release of zoospores through dehiscence tubes or into semi-persistent or persistent vesicles clearly differentiate Halophytophthora from the other two genera (Ho & Jong 1990, Nakagiri et al. 2001, Yang & Hong 2014). In Nothophytophthora, morphological structures are on average smaller than in most Phytophthora and Halophytophthora species but the ranges overlap widely. A significant difference between Nothophytophthora and Phytophthora is the presence of a conspicuous, opaque plug inside the sporangiophore close to the base of most mature sporangia in all known Nothophytophthora species which enables partial caducity in several species including N. caduca, N. chlamydospora, N. valdiviana and N. vietnamensis. A similar plug is also found in many Halophytophthora species (Ho & Jong 1990, Nakagiri et al. 2001). Also, intraspecific co-occurrence of caducity and non-papillate sporangia with internal nested and extended proliferation as in N. caduca and N. valdiviana is not known from any Phytophthora species except the recently described hybrid P. xheterohybrida from subtropical monsoon forests in Taiwan (Jung et al. 2017b).
The LSU, Btub, HSP90, cox1 and NADH1 sequences of N. amphigynosa, N. caduca, N. chlamydospora, N. intricata, N. valdiviana and N. vietnamensis contain 31, 53, 29, 7, 31 and 9 unique polymorphisms, respectively, and differ from each other in 19–116 positions (Table 3, 4, 5, 6, 7, 8). In the ITS rDNA region the six Nothophytophthora species had 0–189 unique polymorphisms and differed from each other at 5–356 positions (Table 2, 10). In addition, they can be easily separated from each other by a combination of morphological and physiological characters of which the most discriminating are highlighted in bold in Table 12. Nothophytophthora amphigynosa differs from the other two homothallic species N. intricata and N. vietnamensis by its predominant production of amphigynous antheridia and by having on average considerably larger sporangia (Table 12; Fig. 4, 7, 9). In addition, N. vietnamensis is distinguished from N. amphigynosa by its partial caducity of sporangia, the predominance of elongated oogonia with tapering often curved bases, different colony morphologies on CA, MEA and V8A, markedly slower growth at 15 and 20 °C, and a higher optimum temperature for growth (Table 12; Fig. 4, 9, 10, 11). Nothophytophthora intricata differs from N. amphigynosa by its lower sporangial l/b ratio, larger oogonial dimensions, the frequent occurrence of intricate, intertwining antheridial stalks, different colony morphologies on all four agar media tested, slower growth at all temperatures, and higher optimum temperature for growth (Table 12; Fig. 4, 7, 10, 11). Nothophytophthora vietnamensis can be separated from its sister species N. intricata by having sporangia which are partially caducous and often formed in dense sympodia, considerably smaller and mostly elongated oogonia with tapering bases, different colony morphologies on all four agar media tested, and faster growth between 10 and 25 °C (Table 12; Fig. 7, 9, 10, 11). Nothophytophthora chlamydospora differs from all other five Nothophytophthora species by the production of chlamydospores. It can also be distinguished from its closest relative N. valdiviana by a markedly higher proportion of caducous sporangia, higher sporangial l/b ratio, absence of internal sporangial proliferation, different colony growth patterns on CA and MEA, considerably slower growth at 25 °C, and lower optimum and maximum temperature for growth (Table 12; Fig. 6, 8, 10, 11); and from N. caduca by its higher sporangial l/b ratio, absence of internal sporangial proliferation, production of sporangia in dense sympodia, different colony growth patterns on all four agar media, considerably slower growth at 25 °C, and lower maximum temperature for growth (Table 12; Fig. 5, 6, 10, 11). Nothophytophthora caduca and N. valdiviana can easily be separated by the predominance of ovoid sporangial shapes, considerably higher level of caducity, absence of dense sympodia and faster growth at 20 °C in N. caduca, and by different colony growth patterns on all four agar media (Table 12; Fig. 5, 8, 10, 11). Although the two populations of N. caduca from two different Valdivian rainforest streams had slightly different optimum and maximum temperatures for growth (Fig. 11) and differed in cox1 at 25 positions (Table 6) their morphology was indistinguishable and, hence, they were considered to belong to the same species.
HOSTS AND GEOGRAPHIC DISTRIBUTION
Nothophytophthora amphigynosa was exclusively isolated from a small forest stream running through a planted mature forest of Eucalyptus globulus, Cupressus sempervirens and Quercus spp. close to Sintra, Portugal (N38°47′28.0″ W9°25′28.7″, 163 m above sea level (a.s.l.)) with a humid Atlantic climate. It co-occurred with two aquatic sterile Phytophthora species, P. amnicola and P. chlamydospora. Nothophytophthora intricata was isolated alongside P. xcambivora, P. megasperma and P. plurivora from the rhizosphere of declining mature Aesculus hippocastanum trees in the floodplain of the river Main in Wiesbaden, Germany (N49°59′48.8″ E8°18′3.2″, 84 m a.s.l.). Nothophytophthora vietnamensis was recovered from rhizosphere soil of Castanopsis sp. and Acer campbellii trees with dieback symptoms on the banks of a small stream in a humid, montane monsoon forest at the Fansipan in Vietnam (N22°19′40.2″ E103°46′53.1″, 2 242 m a.s.l.) where it co-occurred with P. attenuata, P. castaneae and P. cinnamomi. Nothophytophthora caduca (S39°58′9.2″ W73°34′13.4″, 198 m a.s.l.; S39°58′17.6″ W73°34′5.3″, 193 m a.s.l.), N. chlamydospora (S39°57′47.2″ W73°37′9.3″, 9 m a.s.l.) and N. valdiviana (S39°58′3.7″ W73°33′41.9″, 175 m a.s.l.) were detected in four forest streams in the Reserva Costera Valdiviana in Chile with the catchments covered by natural Valdivian rainforests and plantations of E. globulus. All three species co-occurred in these streams with P. chlamydospora and P. kernoviae.
DISCUSSION
During various surveys of oomycete diversity in Europe, Chile and Vietnam slow growing cryptic isolates were detected along-side a diverse community of Phytophthora, Pythium and Phytopythium species from rhizosphere soil and streams in natural and semi-natural ecosystems. Phylogenetic analyses of sequences from the nuclear ITS, LSU, Btub and HSP90 genes and the mitochondrial cox1 and NADH1 genes placed them into six distinct previously unknown species belonging to a new genus described here as Nothophytophthora gen. nov. Phylogenetic analyses together with detailed morphological and physiological studies allowed the taxonomic description of these six taxa as N. amphigynosa, N. caduca, N. chlamydospora, N. intricata, N. valdiviana and N. vietnamensis.
Sparrow (1960, 1976) accepted only four orders in the oomycetes, i.e., Leptomitales, Saprolegniales, Lagenidiales and Peronosporales. Dick et al. (1984) and Dick (2001) proposed the order Pythiales. In the latter publications the number of orders in the oomycota was increased to 12 and in early phylogenies (Riethmüller et al. 1999, Cooke et al. 2000) the Pythiales were accepted and Pythium and the Pythiaceae were assigned to this order. However, Pythium and the Pythiaceae were originally assigned to the Peronosporales (Fischer 1892, Schröter 1893). More recently, refined multigene phylogenies have abandoned the order Pythiales and returned to the four orders accepted by Sparrows (1960, 1976). We followed this trend since in the multigene phylogeny of the present study Pythium s.lat. grouped within a larger group that included the Salisapiliaceae and the Peronosporaceae, both considered belonging to the Peronosporales (Hulvey et al. 2010, Thines & Choi 2016). The multigene phylogeny of this work demonstrated that Nothophytophthora and Phytophthora including the downy mildews form a monophyletic clade with Halophytophthora s.str. clustering basal to this clade and Phytopythium residing in a basal position of this group which forms the Peronosporaceae, order Peronosporales, sensu Baxter et al. (2010), Hulvey et al. (2010) and Thines & Choi (2016). Halophytophthora operculata resided in a basal position to the genus Phytopythium supporting earlier suggestions that this species should be transferred to Phytopythium (Marano et al. 2014, De Cock et al. 2015). In contrast, H. epistomium clearly belongs to a new genus outside of both the Peronosporaceae and Pythiaceae. The six Nothophytophthora species showed nucleotide sequence similarities to the related genera Phytophthora (P. boehmeriae, P. humicola and P. rubi), Halophytophthora (H. avicenniae) and Phytopythium (Ph. helicoides) of 90.8–92.1 %, 90.8–91.0 % and 88.1–88.6 % across the five coding genes LSU, Btub, HSP90, cox1 and NADH1, and of 56.8–68.1 %, 53.9–63.7 % and 46.8–58.5 % in the ITS region.
Despite the high number of oomycete surveys performed during the previous two decades in both managed and natural ecosystems, only four GenBank entries match Nothophytophthora. Isolates PR13-109 and PR12-475 (GenBank accessions KT633938 and KT633937) obtained in 2014 and 2015 from streams in Ireland and isolate REB326-69 (JX122744) from a stream in New Zealand, all originally designated as Phytophthora sp. (Table 1; Than et al. 2013, O’Hanlon et al. 2016). Unfortunately, only ITS sequences were available for these Nothophytophthora isolates preventing their inclusion in the multigene phylogenetic analyses of the present study. However, in a phylogenetic analysis of ITS sequences the three isolates resided in a clade formed by N. chlamydospora and N. valdiviana from Chile with the Irish isolate PR12-475 being basal to the clade and isolates PR13-109 and REB326-69 clustering in a non-supported sister position to N. valdiviana. The relatedness of these congeneric isolates to N. chlamydospora is also demonstrated by the production of chlamydospores by both Irish isolates (Richard O’Hanlon, pers. comm.). In a metagenomic stream survey in the Spanish Pyrenees, Català et al. (2015) found a phylotype (‘MOTU 33’) which resided in Nothophytophthora in a separate phylogenetic analysis of shorter ITS sequences (data not shown). In a yet unpublished metagenomic stream survey in Scotland another Nothophytophthora phylotype was found (David E.L. Cooke, pers. comm.). The findings of Nothophytophthora isolates and phylotypes in Germany, Portugal, Ireland, Scotland, Spain, Chile, New Zealand and Vietnam indicate a wide distribution of this new genus. The scarcity of records in GenBank is most likely caused by the slow growth of Nothophytophthora species which hampers their isolation in presence of faster growing oomycetes like Pythium s.lat., Phytopythium and Phytophthora.
Multiple heterozygous positions in the ITS and LSU sequences of N. amphigynosa, N. chlamydospora and N. vietnamensis, in the HSP90 sequences of N. caduca and N. chlamydospora, and in particular in the Btub sequence of N. valdiviana might indicate reticulation events. In the genus Phytophthora, several studies have demonstrated that interspecific hybridisations play a major evolutionary role by facilitating adaptation to new environments and expansion of host ranges (Brasier et al. 2004, Man in’ t Veld et al. 2012, Bertier et al. 2013, Burgess 2015, Husson et al. 2015, Jung et al. 2017a, b). Also autopolyploidisations and whole genome duplications are suggested having contributed to the evolutionary and pathogenic success of Phytophthora species (Sansome & Brasier 1974, Sansome 1977, Martens & Van de Peer 2010). Determination of nuclear genome sizes and ploidy levels using flow cytometry or genotyping-by-sequencing, and cloning and sequence analyses of single-copy nuclear genes are required to clarify whether and which of the six Nothophytophthora species originate from interspecific hybridisations or autopolyploidisations (Martens & Van de Peer 2010, Bertier et al. 2013, Burgess 2015, Jung et al. 2017b). Interestingly, the two populations of N. caduca have identical NADH1 sequences but differ in their cox1 sequences at 25 positions. This is similar to what has been observed in the three allopolyploid hybrid species P. xcambivora, P. xincrassata and P. xheterohybrida and might be explained by paternal leakage occurring during an interspecific hybridisation event (Jung et al. 2017b) which has been demonstrated as a feasible pathway by which mtDNA might become non-clonal (Eyre-Walker & Awadalla 2001).
Despite being either homothallic or self-sterile, five of the six Nothophytophthora species were able to stimulate oogonia production in an A2 tester strain of P. cinnamomi, a phenomenon also common in homothallic and self-sterile Phytophthora species (Erwin & Ribeiro 1996, Brasier et al. 2003, Jung et al. 2011). Consequently, both genera most likely share the same A1/A2 compatibility system. Traits shared between closely related taxa or groups of taxa were most likely already present in their common ancestor (Yokohama 2002, Carroll 2009, Baum & Smith 2012). Therefore, comparisons of morphological structures of Nothophytophthora and Phytophthora allow clues about the potential morphology and ecology of their common ancestor for which the provisional name ‘Protophytophthora’ is suggested. ‘Protophytophthora’ most likely had a heterothallic A1/A2 breeding system, smooth-walled oogonia with both amphigynous and in the case of selfing also paragynous antheridia, non-papillate sporangia which released already differentiated zoospores without dehiscence tube and proliferated externally and internally in a nested and extended way. Presence of a plug of wall material above the septum where sporangia are being shed and uniform pedicel length are considered as main criteria for true caducity of sporangia in Phytophthora (Al-Hedaithy & Tsao 1979a, b, Erwin & Ribeiro 1996). Since sporangia of N. caduca, N. chlamydospora, N. valdiviana and N. vietnamensis exclusively break off at the base of particularly big opaque plugs of uniform size formed inside the sporangiophore close to the sporangial base, these species should be contemplated as truly caducous and partially airborne species. However, in contrast to airborne species from Phytophthora clades 1–4, 8 and 10, no separate pedicel is formed between the plug and the sporangial base. It can, therefore, be inferred that caducity of sporangia in both genera most likely evolved separately in a convergent way and that their common ancestor ‘Protophytophthora’ most likely had persistent sporangia indicating a soil- and/or waterborne lifestyle.
Four of the six Nothophytophthora species, N. amphigynosa, N. caduca, N. chlamydospora and N. valdiviana, and the Nothophytophthora isolates from Ireland (O’Hanlon et al. 2016) and New Zealand (Than et al. 2013) originated from river systems while N. intricata and N. vietnamensis were isolated from rhizosphere soil of riparian forests. Even though this might indicate an aquatic lifestyle, major morphological and physiological features of these Nothophytophthora species are differing from typical aquatic oomycetes, in particular from the genus Phytophthora. The majority of predominantly aquatic Phytophthora species is characterised by persistent sporangia with abundant nested and extended, internal proliferation, a sterile or disrupted breeding system, fast growth and high optimum and maximum temperatures for growth, all considered specific adaptations to an aquatic lifestyle as saprophytes and opportunistic pathogens (Brasier et al. 2003, Jung et al. 2011, 2017a, Yang & Hong 2013, Yang et al. 2014). In contrast, all known Nothophytophthora species show very slow growth and low maximum temperatures for growth disqualifying them as competitive colonisers of leaf litter. In addition, N. amphigynosa, N. intricata and N. vietnamensis are homothallic and together with N. chlamydospora lack internal proliferation. Moreover, caducity of sporangia as in N. caduca, N. chlamydospora, N. valdiviana and N. vietnamensis has never been observed in predominantly aquatic or in any saprophytic oomycete species. Therefore, it seems possible that their inoculum in the forest streams resulted from canopy drip and surface water flows rather than indicating an aquatic lifestyle. This was also recently suggested for the occurrence of P. xheterohybrida, a hybrid species with functional heterothallic breeding system and partially caducous sporangia, in Taiwanese forest streams (Jung et al. 2017b).
Besides Nothophytophthora and Phytophthora, the only oomycetes with caducous sporangia are the white rusts from the genera Albugo, Pustula and Wilsoniana (Albuginaceae) and the 19 downy mildew genera including Bremia, Graminivora, Hyaloperonospora, Peronospora, Plasmopara, Sclerophthora and Viennotia (Peronosporaceae) (Thines & Choi 2016). White rusts and downy mildews are exclusively obligate biotrophic whereas all airborne and partially airborne Phytophthora species from Clades 1–4, 8 and 10 are facultative, hemibiotrophic or necrotrophic pathogens (Erwin & Ribeiro 1996, Thines & Choi 2016). Since to date no airborne saprophytic oomycetes are known the partial caducity of sporangia in N. caduca, N. chlamydospora, N. valdiviana and N. vietnamensis suggests that these four partially airborne species are most likely facultative, hemibiotrophic or necrotrophic pathogens. Partial caducity of sporangia with varying proportions of caducity between different isolates of the same species also occurs in several Phytophthora species, including P. pseudosyringae and P. psychrophila (Jung et al. 2002, 2003) providing these pathogens with flexible pathogenic opportunities. Both Phytophthora species cause fine root losses in several tree species (Jung et al. 2002, 2003, Jung 2009, Pérez-Sierra et al. 2013). In addition, P. pseudosyringae causes leaf necroses and shoot dieback in Hedera helix and Vaccinium myrtillus, collar rot in Alnus glutinosa, Castanea sativa and Fagus sylvatica and aerial bark cankers on F. sylvatica, Nothofagus alpina, Nothofagus obliqua and Notholithocarpus densiflora (Jung et al. 2003, 2013, Jung & Blaschke 2004, Beales et al. 2009, Jung 2009, Scanu et al. 2010, 2014b, Hansen et al. 2012, Scanu & Webber 2016). In order to clarify whether a similarly flexible pathogenic lifestyle also applies to the four partially caducous Nothophytophthora species, surveys of both root symptoms and above-ground symptoms like wilting of leaves and shoots, leaf and fruit blights and aerial bark cankers for presence of Nothophytophthora species in Valdivian rainforests and Vietnamese mountain forests are needed. Presence of tree dieback in all forests from which the six Nothophytophthora spp. were recovered also suggests that they may have a pathogenic rather than a saprophytic lifestyle. However, pathogenicity tests are urgently required to clarify this. In addition, molecular studies are currently underway to examine whether the six Nothophytophthora species produce elicitins like many Phytophthora species. Since elicitins constitute a group of small proteins involved as effectors in pathogenesis of Phytophthora spp. (Derevnina et al. 2016), their presence in Nothophytophthora would strongly indicate a pathogenic lifestyle for members of this new genus.
Three Nothophytophthora species, N. caduca, N. chlamydospora and N. valdiviana, were recovered from four streams within a few kilometres range in natural Valdivian rainforests in the Reserva Costera Valdiviana. In both the 3-genes (LSU-ITS-cox1) and the 6-genes (LSU-ITS-Btub-HSP90-cox1-NADH1) phylogenetic analyses N. chlamydospora and N. valdiviana constituted sister species. In the 3-gene-analysis they formed a Chilean cluster with N. caduca whereas in the 6-gene-analysis the latter species resided in a basal position of the genus. The two populations of N. caduca from different streams clustered in both analyses separately due to differences of 25 bp in their cox1 sequences. These results strongly indicate that these three Nothophytophthora species are endemic resulting from a sympatric species radiation in the Valdivian rainforests. However, it cannot be excluded that they were introduced with infested nursery stock used in the Eucalyptus globulus and Pinus radiata plantations established in the area of the Reserva Costera Valdiviana in previous decades before the park obtained protection status. Nothophytophthora vietnamensis appears to be native to the mountain forests at the Fansipan due to their remote location and absence of any planting activities. The production of caducous sporangia by N. caduca, N. chlamydospora, N. valdiviana and N. vietnamensis as adaptation to a partially aerial lifestyle in these humid habitats with long term average annual precipitations of approximately 2 500 mm in Valdivia and 2 763 mm in Hoàng Liên Nationalpark, respectively (https://en.wikipedia.org/wiki/Valdivia#Climate; https://en.wikipedia.org/wiki/Sa_Pa#Climate), supports the endemism hypothesis. Interestingly, the two Nothophytophthora species with exclusively persistent sporangia, N. amphigynosa and N. intricata which is the closest relative of N. vietnamensis, were isolated from regions with considerably drier climates less suitable for aerial sporangial production, in Sintra, Portugal (849 mm average annual precipitation) and Wiesbaden, Germany (562 mm) (https://en.climate-data.org).
Some of the most important questions in current oomycete research relate to the evolutionary history and the divergence times of different genera and phylogenetic clades, and how these relate to biogeographical data and the potential centres of origin of invasive pathogens. Using three distinct Bayesian molecular clock models on complete genome sequences of a representative range of oomycete genera, diatoms and a brown algae species, Matari & Blair (2014) estimated the divergence time of the major phylogenetic clades of Phytophthora at 19.8–39.0 million years (Myr) ago. However, such an evolutionary young age does not correspond to recent insights into the natural biogeography of the genus Phytophthora. An accumulating body of indirect evidence resulting from numerous Phytophthora surveys in natural, semi-natural and managed ecosystems across different continents, host range studies, various whole-genus phylogenetic studies and population genetic studies of several globally distributed Phytophthora species is suggesting for the genus Phytophthora two main centres of origin in Southeast Asia and South- and Central America and secondary centres in Western Australia, Europe and North America (Goodwin 1997, Hansen et al. 2000, 2012, 2017, Jung et al. 2002, 2003, 2011, 2016, 2017a, b, Zeng et al. 2009, Rea et al. 2011, Reeser et al. 2011, Vettraino et al. 2011, Blair et al. 2012, Brasier et al. 2012, Kroon et al. 2012, Huai et al. 2013, Oh et al. 2013, Martin et al. 2014, Scanu et al. 2014a, b, Burgess et al. 2017, Arentz 2017). This biogeographical pattern suggests that the genus Phytophthora was already existing before the separation of Gondwana and Laurasia c. 210–175 Myr ago. The considerable discrepancy between an age of 19.8–39 Myr estimated by the molecular clocks and the minimum age suggested by biogeography and plate tectonics might be explained by problems arising from the necessity of calibrating molecular clocks using fossil records. The low preservation potential of oomycetes in combination with the similarity of structures of primitive oomycetes to those from various fungal groups including Zygomycetes, Chytridiomycetes and other zoosporic true fungi make a reliably identified fossil record extremely difficult (Krings et al. 2011). Due to the generally poor microfossil record it is likely that much older oomycete fossils are still awaiting their detection which would change the average evolutionary rates and, hence, the divergence times considerably. Including Nothophytophthora in future coalescence analyses might help to date the divergence time between Phytophthora and Nothophytophthora and give new insights into the evolutionary history of Phytophthora.
Acknowledgments
The authors are grateful to the Portuguese Science and Technology Foundation (FCT) for co-financing with Portuguese national funds the European BiodivERsA project RESIPATH: Responses of European Forests and Society to Invasive Pathogens (BIODIVERSA/0002/2012), and to the Czech Ministry for Education, Youth and Sports and the European Regional Development Fund for financing the Project Phytophthora Research Centre Reg. No. CZ.02.1.01/0.0/0.0/15_003/0000453. Fieldwork in Portugal had logistic support from the Institute for the Conservation of Nature and Forestry (ICNF). DNA sequencing in this study was partly supported by the Hungarian Scientific Research Fund (OTKA) grant K101914 and by grant MIUR-FIRB 2010 (RBFR10PZ4N) from the Italian Ministry of Education, University and Research (MIUR) ‘Metagenomic strategies to assess genetic diversity in soil-borne Phytophthora species’. This work has also received funding from the European Union’s Horizon 2020 research and innovation programme under grant agreement No. 635646, POnTE (Pest Organisms Threatening Europe).
REFERENCES
- Al-Hedaithy SSA, Tsao PH. 1979a. The effects of culture media and sporulation methods on caducity and pedicel length of sporangia in selected species of Phytophthora. Mycologia 71: 392–401. [Google Scholar]
- Al-Hedaithy SSA, Tsao PH. 1979b. Sporangium pedicel length in Phytophthora species and the consideration of its uniformity in determining sporangium caducity. Transactions of the British Mycological Society 72: 1–13. [Google Scholar]
- Arentz F. 2017. Phytophthora cinnamomi A1: An ancient resident of New Guinea and Australia of Gondwanan origin? Forest Pathology, in press. doi: 10.1111/efp.12342.
- Bala K, Robideau GP, Lévesque CA, et al. 2010. Phytopythium gen. nov. and Phytopythium sindhum sp. nov. Fungal Planet 49. Persoonia 24: 136–137. [Google Scholar]
- Balci Y, Balci S, Eggers J, et al. 2007. Phytophthora spp. associated with forest soils in Eastern and North-Central U.S. oak ecosystems. Plant Disease 91: 705–710. [DOI] [PubMed] [Google Scholar]
- Balci Y, Halmschlager E. 2003a. Incidence of Phytophthora species in oak forests in Austria and their possible involvement in oak decline. Forest Pathology 33: 157–174. [Google Scholar]
- Balci Y, Halmschlager E. 2003b. Phytophthora species in oak ecosystems in Turkey and their association with declining oak trees. Plant Pathology 52: 694–702. [Google Scholar]
- Baum DA, Smith SD. 2012. Tree thinking: an introduction to phylogenetic biology. Roberts & Co., Greenwood Village, Colorado. [Google Scholar]
- Baxter L, Tripathy S, Ishaque L, et al. 2010. Signatures of adaptation to obligate biotrophy in the Hyaloperonospora arabidopsis genome. Science 330: 1549–1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beakes GW, Glockling SL, Sekimoto S. 2012. The evolutionary phylogeny of the oomycete “fungi”. Protoplasma 249: 3–19. [DOI] [PubMed] [Google Scholar]
- Beakes GW, Honda T, Thines M. 2014. Systematics of the Stramenipila: Labyrinthulomycota, Hyphochytridiomycota, and Oomycota. In: McLaughlin DJ, Spatafora J. (eds), Systematics and evolution: 39–97. Springer, New York. [Google Scholar]
- Beales P, Giltrap PM, Webb KM, et al. 2009. A further threat to UK heathland bilberry Vaccinium myrtillus by Phytophthora pseudosyringae. Plant Pathology 59: 406. [Google Scholar]
- Bertier L, Leus L, D’hondt L, et al. 2013. Host adaptation and speciation through hybridization and polyploidy in Phytophthora. PloS ONE 8: e85385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blair JE, Coffey MD, Martin FN. 2012. Species tree estimation for the late blight pathogen, Phytophthora infestans, and close relatives. PLoS ONE 7: e37003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blair JE, Coffey MD, Park S-Y, et al. 2008. A multi-locus phylogeny for Phytophthora utilizing markers derived from complete genome sequences. Fungal Genetics and Biology 45: 266–277. [DOI] [PubMed] [Google Scholar]
- Brasier CM. 1967. Physiology of reproduction in Phytophthora. PhD thesis, University of Hull, UK. [Google Scholar]
- Brasier CM, Cooke DEL, Duncan JM, et al. 2003. Multiple new phenotypic taxa from trees and riparian ecosystems in Phytophthora gonapodyides – P. megasperma ITS Clade 6, which tend to be high-temperature tolerant and either inbreeding or sterile. Mycological Research 107: 277–290. [DOI] [PubMed] [Google Scholar]
- Brasier CM, Franceschini S, Vettraino AM, et al. 2012. Four phenotypically and phylogenetically distinct lineages in Phytophthora lateralis. Fungal Biology 116: 1232–1249. [DOI] [PubMed] [Google Scholar]
- Brasier CM, Kirk SA, Delcan J, et al. 2004. Phytophthora alni sp. nov. and its variants: designation of emerging heteroploid hybrid pathogens spreading on Alnus trees. Mycological Research 108: 1172–1184. [DOI] [PubMed] [Google Scholar]
- Brasier CM, Robredo F, Ferraz JFP. 1993. Evidence for Phytophthora cinnamomi involvement in Iberian oak decline. Plant Pathology 42: 140–145. [Google Scholar]
- Brasier CM, Webber J. 2010. Sudden larch death. Nature 466: 824–825. [DOI] [PubMed] [Google Scholar]
- Briard M, Dutertre M, Rouxel F, et al. 1995. Ribosomal RNA sequence divergence within the Pythiaceae. Mycological Research 99: 1119–1127. [Google Scholar]
- Burgess TI. 2015. Molecular characterization of natural hybrids formed between five related indigenous Clade 6 Phytophthora species. PLoS ONE 10: e0134225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess TI, White D, McDougall KM, et al. 2017. Distribution and diversity of Phytophthora across Australia. Pacific Conservation Biology 23: 1–13. [Google Scholar]
- Carroll SB. 2009. The making of the fittest: DNA and the ultimate forensic record of evolution. Quercus Publishing, London, UK. [Google Scholar]
- Català S, Peréz-Sierra A, Abad-Campos P. 2015. The use of genus-specific amplicon pyrosequencing to assess Phytophthora species diversity using eDNA from soil and water in Northern Spain. PLoS ONE 10: e0119311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooke DEL, Drenth A, Duncan JM, et al. 2000. A molecular phylogeny of Phytophthora and related oomycetes. Fungal Genetics and Biology 30: 17–32. [DOI] [PubMed] [Google Scholar]
- De Cock AWAM, Lévesque CA. 2004. New species of Pythium and Phytophthora. Studies in Mycology 50: 481–487. [Google Scholar]
- De Cock AWAM, Lodhi AM, Rintoul TL, et al. 2015. Phytopythium: molecular phylogeny and systematics. Persoonia 34: 25–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derevnina L, Dagdas YF, De la Concepcion JC, et al. 2016. Nine things to know about elicitins. New Phytologist 212: 888–895. [DOI] [PubMed] [Google Scholar]
- Dick MW. 1990. Keys to Pythium. University of Reading Press, Reading, UK. [Google Scholar]
- Dick MW. 2001. Straminipilous fungi: systematics of the Peronosporomycetes including accounts of the marine straminipilous protists, the plasmodiophorids and similar organisms. Kluwer Academic Publishers, Dordrecht, The Netherlands. [Google Scholar]
- Dick MW, Wong PTW, Clark G. 1984. The identity of the oomycete causing ‘kikuyu yellows’ with a reclassification of the downy mildews. Botanical Journal of the Linnean Society 89: 171–198. [Google Scholar]
- Erwin DC, Ribeiro OK. 1996. Phytophthora diseases worldwide. APS Press, St. Paul, Minnesota. [Google Scholar]
- Eyre-Walker A, Awadalla P. 2001. Does human mtDNA recombine? Journal of Molecular Evolution 53: 430–435. [DOI] [PubMed] [Google Scholar]
- Fischer A. 1892. Rabenhorst’s Kryptogamen-Flora, Pilze - Phycomycetes. 2nd edition, Eduard Kummer, Leipzig. [Google Scholar]
- Gallegly ME, Hong C. 2008. Phytophthora: identifying species by morphology and DNA fingerprints. APS Press, St. Paul, Minnesota. [Google Scholar]
- Gäumann EA. 1952. The fungi. A description of their morphological features and evolutionary development. Hafner Publishing, New York & London. [Google Scholar]
- Ginetti B, Moricca S, Squires JN, et al. 2014. Phytophthora acerina sp. nov., a new species causing bleeding cankers and dieback of Acer pseudoplatanus trees in planted forests in Northern Italy. Plant Pathology 63: 858–876. [Google Scholar]
- Göker M, Voglmayer H, Riethmüller A, et al. 2007. How do obligate parasites evolve? A multi-gene phylogenetic analysis of downy mildews. Fungal Genetics and Biology 44: 105–122. [DOI] [PubMed] [Google Scholar]
- Goodwin SB. 1997. The population genetics of Phytophthora. Phytopathology 87: 462–473. [DOI] [PubMed] [Google Scholar]
- Green S, Brasier CM, Schlenzig A, et al. 2013. The destructive invasive pathogen Phytophthora lateralis found on Chamaecyparis lawsoniana across the UK. Forest Pathology 43: 19–28. [Google Scholar]
- Green S, Elliot M, Armstrong A, et al. 2015. Phytophthora austrocedrae emerges as a serious threat to juniper (Juniperus communis) in Britain. Plant Pathology 64: 456–466. [Google Scholar]
- Greslebin A, Hansen EM, Sutton W. 2007. Phytophthora austrocedrae sp. nov., a new species associated with Austrocedrus chilensis mortality in Patagonia Argentina. Mycological Research 111: 308–316. [DOI] [PubMed] [Google Scholar]
- Hansen EM, Goheen DJ, Jules ES, et al. 2000. Managing Port-Orford-Cedar and the introduced pathogen Phytophthora lateralis. Plant Disease 84: 4–14. [DOI] [PubMed] [Google Scholar]
- Hansen EM, Reeser PW, Sutton W. 2012. Phytophthora beyond agriculture. Annual Review of Phytopathology 50: 359–378. [DOI] [PubMed] [Google Scholar]
- Hansen EM, Reeser PW, Sutton W. 2017. Ecology and pathology of Phytophthora ITS clade 3 species in forests in western Oregon, USA. Mycologia 109: 100–114. [DOI] [PubMed] [Google Scholar]
- Hardham AR. 2005. Phytophthora cinnamomi. Molecular Plant Pathology 6: 589–604. [DOI] [PubMed] [Google Scholar]
- Henricot B, Pérez-Sierra A, Jung T. 2014. Phytophthora pachypleura sp. nov., a new species causing root rot of Aucuba japonica and other ornamentals in the United Kingdom. Plant Pathology 63: 1095–1109. [Google Scholar]
- Ho HH, Jong SC. 1990. Halophytophthora, gen. nov., a new member of the family Pythiaceae. Mycotaxon 36: 377–382. [Google Scholar]
- Huai WX, Tian G, Hansen EM, et al. 2013. Identification of Phytophthora species baited and isolated from forest soil and streams in northwestern Yunnan province, China. Forest Pathology 43: 87–103. [Google Scholar]
- Hüberli D, Hardy GEStJ, White D, et al. 2013. Fishing for Phytophthora from Western Australia’s waterways: A distribution and diversity survey. Australasian Plant Pathology 42: 251–260. [Google Scholar]
- Huelsenbeck JP, Ronquist F. 2001. MrBayes: Bayesian inference of phylogenetic trees. Bioinformatics 17: 754–755. [DOI] [PubMed] [Google Scholar]
- Hulvey J, Telle S, Nigrelli L, et al. 2010. Salisapiliaceae – a new family of oomycetes from marsh grass litter of southeastern North America. Persoonia 25: 109–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husson C, Aguayo J, Revellin C, et al. 2015. Evidence for homoploid speciation in Phytophthora alni supports taxonomic reclassification in this species complex. Fungal Genetics and Biology 77: 12–21. [DOI] [PubMed] [Google Scholar]
- Jung T. 2009. Beech decline in Central Europe driven by the interaction between Phytophthora infections and climatic extremes. Forest Pathology 39: 73–94. [Google Scholar]
- Jung T, Blaschke H, Neumann P. 1996. Isolation, identification and pathogenicity of Phytophthora species from declining oak stands. European Journal of Forest Pathology 26: 253–272. [Google Scholar]
- Jung T, Blaschke H, Osswald W. 2000. Involvement of soilborne Phytophthora species in Central European oak decline and the effect of site factors on the disease. Plant Pathology 49: 706–718. [Google Scholar]
- Jung T, Blaschke M. 2004. Phytophthora root and collar rot of alders in Bavaria: distribution, modes of spread and possible management strategies. Plant Pathology 53: 197–208. [Google Scholar]
- Jung T, Burgess TI. 2009. Re-evaluation of Phytophthora citricola isolates from multiple woody hosts in Europe and North America reveals a new species, Phytophthora plurivora sp. nov. Persoonia 22: 95–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung T, Chang TT, Bakonyi J, et al. 2017a. Diversity of Phytophthora species in natural ecosystems of Taiwan and association with disease symptoms. Plant Pathology 66: 194–211. [Google Scholar]
- Jung T, Cooke DEL, Blaschke H, et al. 1999. Phytophthora quercina sp. nov., causing root rot of European oaks. Mycological Research 103: 785–798. [Google Scholar]
- Jung T, Hansen EM, Winton L, et al. 2002. Three new species of Phytophthora from European oak forests. Mycological Research 106: 397–411. [Google Scholar]
- Jung T, Horta Jung M, Scanu B, et al. 2017b. Six new Phytophthora species from ITS Clade 7a including two sexually functional heterothallic hybrid species detected in natural ecosystems in Taiwan. Persoonia 38: 100–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung T, Nechwatal J, Cooke DEL, et al. 2003. Phytophthora pseudosyringae sp. nov., a new species causing root and collar rot of deciduous tree species in Europe. Mycological Research 107: 772–789. [DOI] [PubMed] [Google Scholar]
- Jung T, Orlikowski L, Henricot B, et al. 2016. Widespread Phytophthora infestations in European nurseries put forest, semi-natural and horticultural ecosystems at high risk of Phytophthora diseases. Forest Pathology 46: 134–163. [Google Scholar]
- Jung T, Stukely MJC, Hardy GEStJ, et al. 2011. Multiple new Phytophthora species from ITS Clade 6 associated with natural ecosystems in Australia: evolutionary and ecological implications. Persoonia 26: 13–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung T, Vettraino AM, Cech TL, et al. 2013. The impact of invasive Phytophthora species on European forests. In: Lamour K. (ed), Phytophthora: a global perspective: 146–158. CABI, Wallingford, UK. [Google Scholar]
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution 30: 772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko WH, Chang HS, Su HJ. 1978. Isolates from Phytophthora cinnamomi from Taiwan as evidence for an Asian origin of the species. Transactions of the British Mycological Society 71: 496–499. [Google Scholar]
- Krings M, Taylor TN, Dotzler N. 2011. The fossil record of the Peronosporomycetes Oomycota. Mycologia 103: 445–457. [DOI] [PubMed] [Google Scholar]
- Kroon LPNM, Bakker FT, Van den Bosch GBM, et al. 2004. Phylogenetic analysis of Phytophthora species based on mitochondrial and nuclear DNA sequences. Fungal Genetics and Biology 41: 766–782. [DOI] [PubMed] [Google Scholar]
- Kroon LPNM, Brouwer H, De Cock AWAM, et al. 2012. The genus Phytophthora anno 2012. Phytopathology 102: 348–364. [DOI] [PubMed] [Google Scholar]
- Man in ’t Veld WA, Rosendahl KCHM, Hong C. 2012. Phytophthora × serendipita sp. nov. and P. × pelgrandis, two destructive pathogens generated by natural hybridization. Mycologia 104: 1390–1396. [DOI] [PubMed] [Google Scholar]
- Marano AV, Jesus AL, De Souza JI, et al. 2014. A new combination in Phytopythium: P. kandeliae (Oomycetes, Straminipila). Mycosphere 5: 510–522. [Google Scholar]
- Martens C, Van de Peer Y. 2010. The hidden duplication past of the plant pathogen Phytophthora and its consequences for infection. BMC Genomics 11: 353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin FN, Blair JE, Coffey MD. 2014. A combined mitochondrial and nuclear multilocus phylogeny of the genus Phytophthora. Fungal Genetics and Biology 66: 19–32. [DOI] [PubMed] [Google Scholar]
- Martin FN, Tooley PW. 2003. Phylogenetic relationships among Phytophthora species inferred from sequence analysis of mitochondrially encoded cytochrome oxidase I and II genes. Mycologia 95: 269–284. [PubMed] [Google Scholar]
- Matari NH, Blair JE. 2014. A multilocus timescale for oomycete evolution estimated under three distinct molecular clock models. BMC Evolutionary Biology 14: 101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moncalvo JM, Wang HH, Hseu RS. 1995. Phylogenetic relationships in Ganoderma inferred from the internal transcribed spacers and 25S ribosomal DNA sequences. Mycologia 87: 223–238. [Google Scholar]
- Nagy LG, Kocsubé S, Csanádi Z, et al. 2012. Re-mind the gap! Insertion – deletion data reveal neglected phylogenetic potential of the nuclear ribosomal internal transcribed spacer ITS of fungi. PLoS ONE 7: e49794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy ZÁ, Bakonyi J, Érsek T. 2003. Standard and Swedish variant types of the hybrid alder Phytophthora attacking alder in Hungary. Pest Management Science 59: 484–492. [DOI] [PubMed] [Google Scholar]
- Nakagiri A. 2001. Ecology and biodiversity of Halophytophthora species. In: Hyde KD, Ho WH, Pointing SB. (eds), Aquatic mycology across the millenium: 153–164. Fungal Diversity Press, Hong Kong. [Google Scholar]
- O’Hanlon R, Choiseul J, Corrigan M, et al. 2016. Diversity and detections of Phytophthora species from trade and nontrade environments in Ireland. Bulletin OEPP/EPPO Bulletin 46: 594–602. [Google Scholar]
- Oh E, Gryzenhout M, Wingfield BD, et al. 2013. Surveys of soil and water reveal a goldmine of Phytophthora diversity in South African natural ecosystems. IMA Fungus 4: 123–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Sierra A, López-García C, León M, et al. 2013. Previously unrecorded low temperature Phytophthora species associated with Quercus decline in a Mediterranean forest in Eastern Spain. Forest Pathology 43: 331–339. [Google Scholar]
- Rea AJ, Burgess TI, Hardy GEStJ, et al. 2011. Two novel and potentially endemic species of Phytophthora associated with episodic dieback of kwongan vegetation in the south-west of Western Australia. Plant Pathology 60: 1055–1068. [Google Scholar]
- Reeser PW, Sutton W, Hansen EM, et al. 2011. Phytophthora species in forest streams in Oregon and Alaska. Mycologia 103: 22–35. [DOI] [PubMed] [Google Scholar]
- Riethmüller A, Voglmayr H, Göker M, et al. 2002. Phylogenetic relationships of the downy mildews Peronosporales and related groups based on nuclear large subunit ribosomal DNA sequences. Mycologia 94: 834–849. [DOI] [PubMed] [Google Scholar]
- Riethmüller A, Weiß M, Oberwinkler F. 1999. Phylogenetic studies of Saprolegniomycetidae and related groups based on nuclear large subunit DNA sequences. Canadian Journal of Botany 77: 1790–1800. [Google Scholar]
- Rizzo DM, Garbelotto M, Davidson JM, et al. 2002. Phytophthora ramorum as the cause of extensive mortality of Quercus spp. and Lithocarpus densiflorus in California. Plant Disease 86: 205–214. [DOI] [PubMed] [Google Scholar]
- Robideau GP, De Cock AWAM, Coffey MD, et al. 2011. DNA barcoding of oomycetes with cytochrome c oxidase subunit I and internal transcribed spacer. Molecular Ecology Resources 11: 1002–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574. [DOI] [PubMed] [Google Scholar]
- Runge F, Telle S, Ploch S, et al. 2011. The inclusion of downy mildews in a multi-locus-dataset and its reanalysis reveals a high degree of paraphyly in Phytophthora. IMA Fungus 2: 163–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sansome ER. 1977. Polyploidy and induced gametangial formation in British isolates of Phytophthora infestans. Journal of General Microbiology 99: 311–316. [Google Scholar]
- Sansome ER, Brasier CM. 1974. Polyploidy associated with varietal differentiation in the megasperma complex of Phytophthora. Transactions of the British Mycological Society 63: 461–467. [Google Scholar]
- Scanu B, Hunter GC, Linaldeddu BT, et al. 2014a. A taxonomic re-evaluation reveals that Phytophthora cinnamomi and P. cinnamomi var. parvispora are separate species. Forest Pathology 44: 1–20. [Google Scholar]
- Scanu B, Linaldeddu BT, Deidda A, et al. 2015. Diversity of Phytophthora species from declining Mediterranean maquis vegetation, including two new species, Phytophthora crassamura and P. ornamentata sp. nov. PLoS ONE 10: e0143234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scanu B, Linaldeddu BT, Franceschini A. 2010. First report of Phytophthora pseudosyringae associated with ink disease of Castanea sativa in Italy. Plant Disease 94: 1068. [DOI] [PubMed] [Google Scholar]
- Scanu B, Linaldeddu BT, Peréz-Sierra A, et al. 2014b. Phytophthora ilicis as a leaf and stem pathogen of Ilex aquifolium in Mediterranean islands. Phytopathologia Mediterranea 53: 480–490. [Google Scholar]
- Scanu B, Webber JF. 2016. Dieback and mortality of Nothofagus in Britain: ecology, pathogenicity and sporulation potential of the causal agent Phytophthora pseudosyringae. Plant Pathology 65: 26–36. [Google Scholar]
- Schröter J. 1893. Die Natürlichen Pflanzenfamilien 11. Engelmann, Leipzig. [Google Scholar]
- Shrestha SK, Zhou Y, Lamour K. 2013. Oomycetes baited from streams in Tennessee 2010–2012. Mycologia 105: 1516–1523. [DOI] [PubMed] [Google Scholar]
- Silvestro D, Michalak I. 2012. raxmlGUI: a graphical front-end for RAxML. Organisms Diversity and Evolution 12: 335–337. [Google Scholar]
- Sparrow FK. 1960. Aquatic Phycomycetes, 2nd revised edition University of Michigan Press, Ann Arbor. [Google Scholar]
- Sparrow FK. 1976. The present status of classification in biflagellate fungi. In: Gareth Jones EB. (ed), Recent Advances in Aquatic Mycology: 213–222. Halsted Press of John Wiley & Sons, NY. [Google Scholar]
- Staden R, Beal KF, Bonfield JK. 2000. The Staden package 1998. Methods in Molecular Biology 132: 115–130. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30: 1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, et al. 2013. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Molecular Biology and Evolution 30: 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Than DJ, Hughes KJD, Boonhan N, et al. 2013. A TaqMan real-time PCR assay for the detection of Phytophthora ‘taxon Agathis’ in soil, pathogen of Kauri in New Zealand. Forest Pathology 43: 324–330. [Google Scholar]
- Thines M, Choi Y-J. 2016. Evolution, diversity and taxonomy of the Peronosporaceae, with focus on the genus Peronospora. Phytopathology 106: 6–18. [DOI] [PubMed] [Google Scholar]
- Thines M, Kamoun S. 2010. Oomycete-plant coevolution: Recent advances and future aspects. Current Opinions in Plant Biology 13: 427–433. [DOI] [PubMed] [Google Scholar]
- Uzuhashi S, Tojo M, Kakishima M. 2010. Phylogeny of the genus Pythium and description of new genera. Mycoscience 51: 337–365. [Google Scholar]
- Vettraino AM, Barzanti GP, Bianco MC, et al. 2002. Occurrence of Phytophthora species in oak stands in Italy and their association with declining oak trees. Forest Pathology 32: 19–28. [Google Scholar]
- Vettraino AM, Brasier CM, Brown AV, et al. 2011. Phytophthora himalsilva sp. nov. an unusually phenotypically variable species from a remote forest in Nepal. Fungal Biology 115: 275–287. [DOI] [PubMed] [Google Scholar]
- Vettraino AM, Morel O, Perlerou C, et al. 2005. Occurrence and distribution of Phytophthora species associated with Ink Disease of chestnut in Europe. European Journal of Plant Pathology 111: 169–180. [Google Scholar]
- Villa NO, Kageyama K, Asano T, et al. 2006. Phylogenetic relationships of Pythium and Phytophthora species based on ITS rDNA, cytochrome oxidase II and beta-tubulin gene sequences. Mycologia 98: 410–422. [DOI] [PubMed] [Google Scholar]
- Voglmayer H. 2003. Phylogenetic relationships of Peronospora and related genera based on nuclear ribosomal ITS sequences. Mycological Research 107: 1132–1142. [DOI] [PubMed] [Google Scholar]
- White TJ, Bruns T, Lee S, et al. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Gelfand DH, Sninsky JJ, Innes MA, et al. (eds), PCR protocols: a guide to methods and applications: 315–322. Academic Press, San Diego, California, USA. [Google Scholar]
- Yang X, Copes WE, Hong C. 2014. Two novel species representing a new clade and cluster of Phytophthora. Fungal Biology 118: 72–82. [DOI] [PubMed] [Google Scholar]
- Yang X, Hong C. 2013. Phytophthora virginiana sp. nov., a high-temperature tolerant species from irrigation water in Virginia. Mycotaxon 126: 167–176. [Google Scholar]
- Yang X, Hong C. 2014. Halophytophthora fluviatilis sp. nov. from freshwater in Virginia. FEMS Microbiological Letters 352: 230–237. [DOI] [PubMed] [Google Scholar]
- Yokohama S. 2002. Molecular evolution of color vision in vertebrates. Gene 300: 69–78. [DOI] [PubMed] [Google Scholar]
- Young ND, Healy J. 2003. GapCoder automates the use of indel characters in phylogenetic analysis. BMC Bioinformatics 4: 6 doi: https://doi.org/10.1186/1471-2105-4-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng H-C, Ho H-H, Zheng F-C. 2009. A survey of Phytophthora species on Hainan Island of South China. Journal of Phytopathology 157: 33–39. [Google Scholar]


