Figure 1.
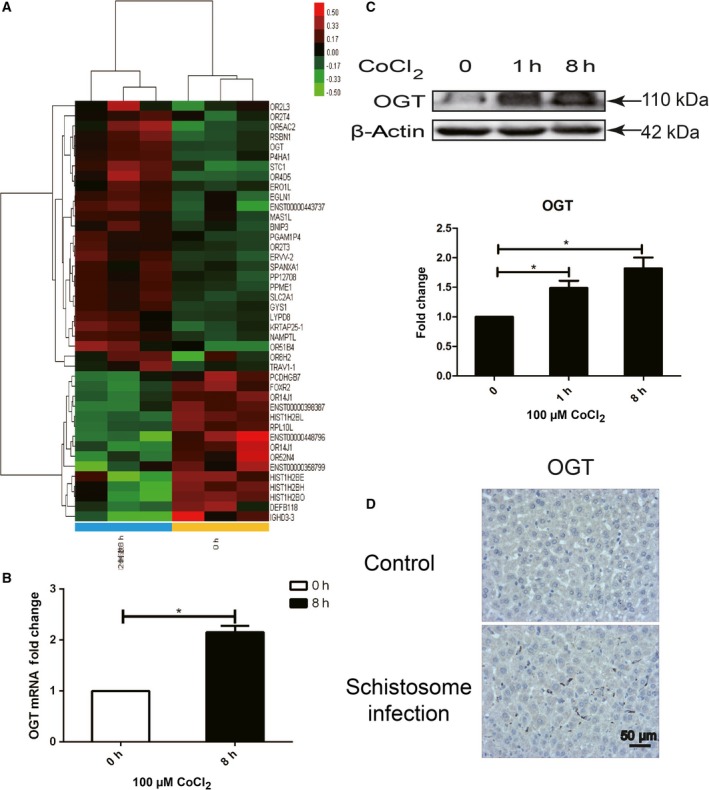
Genes related to histone methylation modification were up‐regulated in hypoxia‐induced LX‐2 cells. Total RNA was extracted and reversely transcribed into cDNA from normal oxygen or CoCl2‐treated hypoxia‐induced human hepatic stellate cell line LX‐2. (A) Cluster analysis of genomewide expression chips (red: up‐regulated genes, green: down‐regulated genes). (B) The expression of Ogt, a representative up‐regulated gene from genomewide expression chips, was detected at mRNA level by qPCR. Densitometric analysis was performed using pooled data from three such experiments. Data were mean ± SD (*P < 0.05). (C) Total protein was extracted at 0, 1, and 8 h from hypoxia‐induced LX‐2 cells, and the expression of OGT was detected by western blot. Densitometric analysis was performed using pooled data from three such experiments. Data were mean ± SD (*P < 0.05). (D) BALB/c female mice, 6–8 weeks old, were percutaneously infected with 25 cercariae of Schistosoma japonicum through the shaved abdomen, sacrificed at 8 weeks postinfection, and samples of liver were collected. The expression of OGT in S. japonicum‐infected (n = 3) and noninfected (n = 3) mice liver was detected with immunohistochemistry and representative images were shown.
