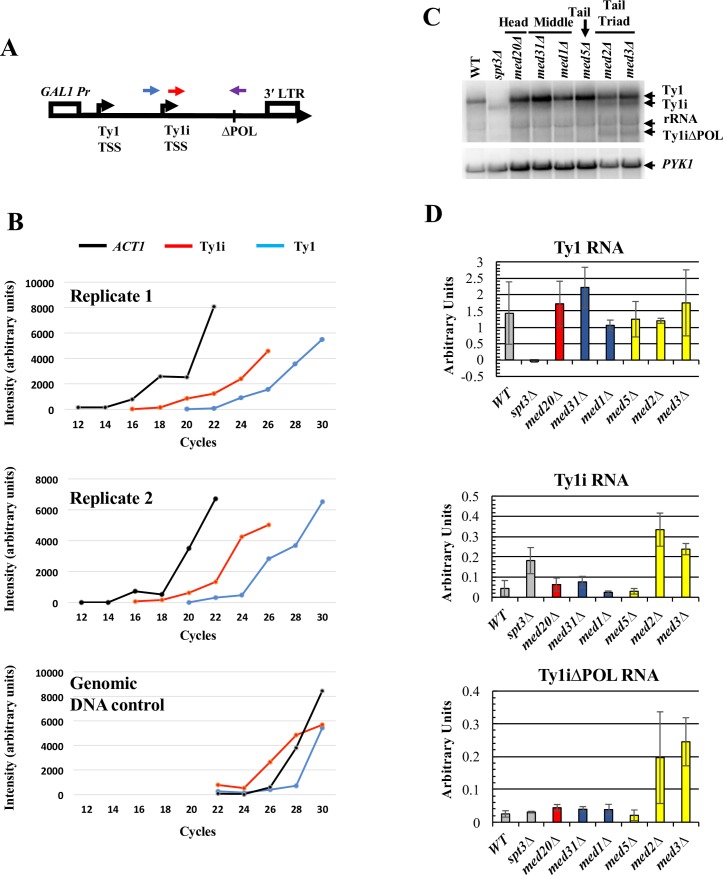
Fig 7. Ty1i situated downstream of a repressed Ty1 element is repressed by Mediator tail module triad subunits.
(A) Schematic of the GAL1:Ty1ΔPOL cassette in pGTy1ΔPOL showing forward primer locations for detection of Ty1 RNA (blue) versus Ty1i RNA (red). Note that because Ty1i is contained within Ty1, the Ty1i primer reports both Ty1i and Ty1 transcripts. Both amplifications utilized the same reverse primer (purple), that crosses the deletion junction and contains sequences unique to the pGTy1ΔPOL element. A reverse primer specific for ACT1 mRNA was also used to synthesize cDNA used as a template for the PCR amplification. No PCR product was detected using the ΔPOL reverse primer when RNA from yeast lacking pGTy1ΔPOL was used as template. (B) Quantitation of the products of Reverse Transciption-PCR reactions using polyA+ RNA isolated from wild-type yeast bearing plasmid pGTy1ΔPOL, and grown in glucose-containing broth, and from a genomic DNA control. Aliquots were taken from reactions at the indicated number of cycles and analyzed by agarose gel electrophoresis (S6 Fig). Levels of RT-PCR amplification products using Ty1, Ty1i, and ACT1 primers are indicated. (C) Northern blot probed with a single-stranded riboprobe that hybridizes to Ty1, Ty1i, Ty1ΔPOL and Ty1iΔPOL RNA. Note that the full-length Ty1ΔPOL transcript cannot be distinguished from the rRNA band. Image is representative of three biological replicates. (D) Quantification for Ty1, Ty1i, and Ty1iΔPOL RNA from three biological replicate northern analyses. Graph bar colors correspond to WT strain and the congenic spt3Δ strain as a negative control (grey bars), Mediator head subunit gene deletion strain (red bars), middle subunit gene deletion strains (blue bars) and tail subunit gene deletion strains (yellow bars). Error bars represent s.d.