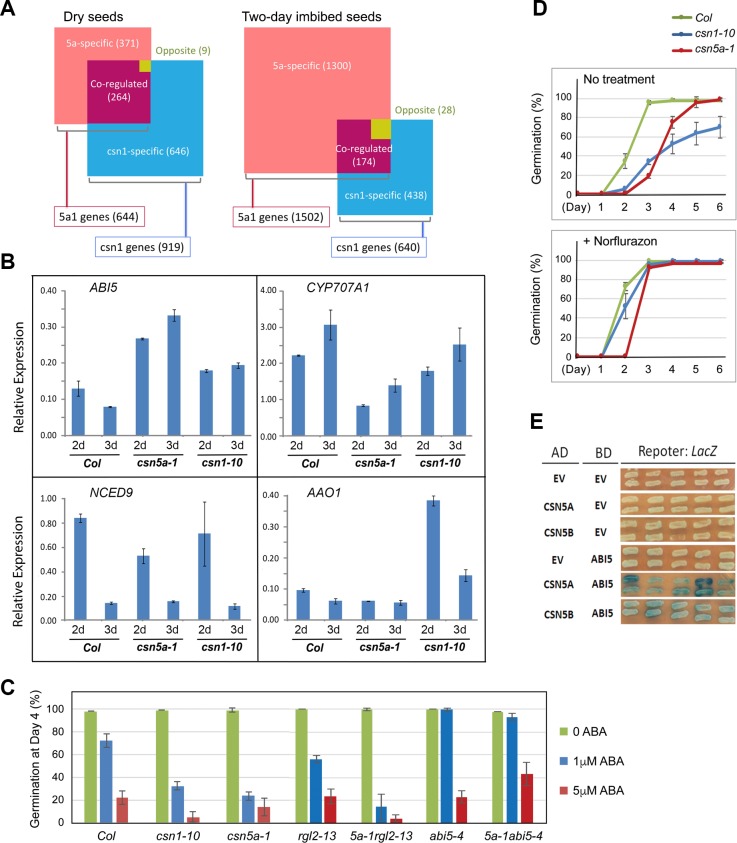
Fig 7. Effects of csn1-10 and csn5a-1 mutations on transcriptome and ABA sensitivity.
(A) The diagrams showing the number of SSTF (statistically significant two-fold change) genes in csn5a-1 and csn1-10 mutants compared to wild type Col in dry seeds, or after two-day imbibition. csn1-10 affected more genes than csn5a-1 in dry seeds, while csn5a-1 affected more genes than csn1-10 in 2-day imbibed seeds. The areas of the squares were drawn in proportion to the number of the genes in the indicated category. (B) Real-time qPCR analysis of representative ABA related genes on day2 and day3 imbibed seeds of Col, csn1-10, and csn5a-1. The expression level of each sample was normalized to IPA-like1 (AT1G17210). (C) Sensitivity to ABA-mediated inhibition of germination is enhanced in csn1-10, csn5a-1, and csn5a-1rgl2 double mutant. The csn5a-1abi5-4 double mutant showed resistance to ABA. Cold stratified seeds were used. Error bars represent standard deviation from 4 repeats. (D) ABA biosynthetic inhibitor norflurazon (5μM) rescued the low germination rates of both csn1-10 and csn5a-1, but did not rescue the delayed germination in csn5a-1. Error bars represent standard deviation from 4 repeats. (E) Yeast-two-hybrid sassy showing interactions of ABI5 with CSN5A and CSN5B. AD, activation-domain fusion vectors; BD, DNA-binding domain fusion vectors; EV, empty vector. See also S6 and S7 Figs.