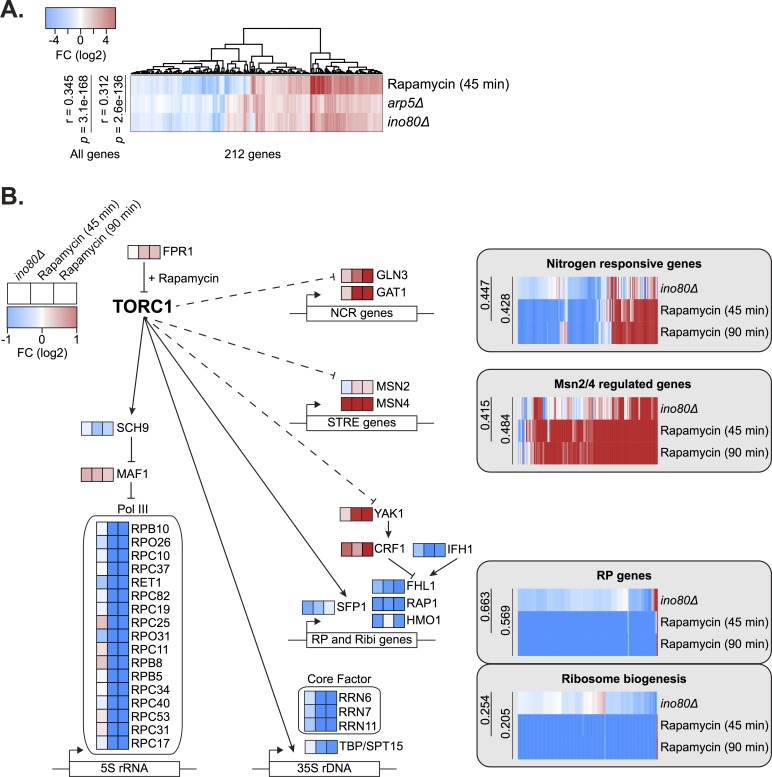
Fig 7. INO80 regulates the expression of key TOR signaling effectors.
(A) Log-transformed Z-scores of expression fold-change (FC) between untreated and treated (30nM rapamycin for 45 minutes) wild-type cells or indicated deletion strains. Genes with at least a 1.5 fold-change are plotted. Pearson correlations and p-values are shown for all genes (>6000) regardless of fold-change difference. (B) Diagram of key genes involved in the TORC1 regulation of nitrogen source quality responsive genes [77], Msn2/4 regulated stress response genes [78], ribosomal protein (RP) genes, and ribosome biogenesis genes [79]. Log-transformed expression fold-change is shown comparing untreated wild-type cells and rapamycin treated (45 and 90 minutes) or deletion strains as indicated. Gene lists are found in S8 Table.