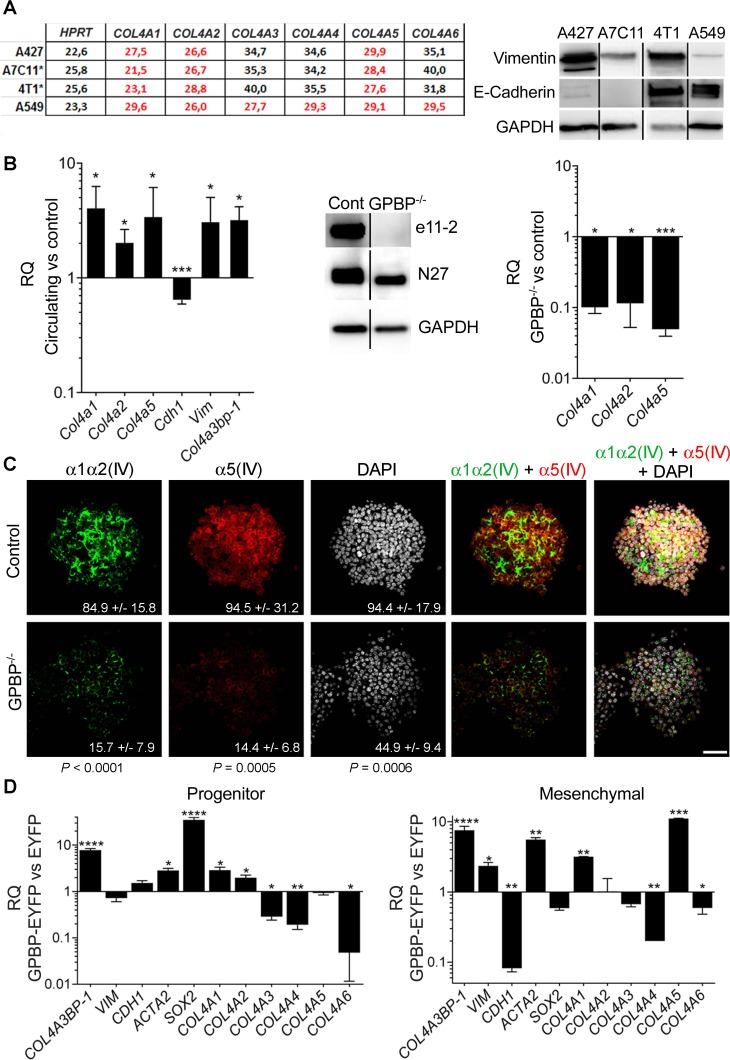
Figure 1. GPBP directs the formation of two independent mesh collagen IV networks in chemoresistant EMT cancer cell phenotypes.
(A) Left, the threshold-cycle (CT) for the indicated mRNA and cell lines was determined by RT-qPCR. Denoted in red characters are predominantly expressed mRNA (*murine breast cancer cell lines). Right, WB analysis of the indicated cell cultures and polypeptides. (B) Left, represented with bars are the relative quantity (RQ) expressed as mean ± standard deviation (SD) of the indicated mRNA in c4T1 cultures (circulating) in comparison (versus, vs) with 4T1 cultures (control). Middle, WB analysis of the indicated 4T1 cell extracts to assess GPBP (e11-2), GPBP and GPBP-2/CERT (N27) and GAPDH (glyceraldehyde 3-phosphate dehydrogenase) expression. Right, RQ of the indicated mRNA and 4T1 cultures is represented as at left. Statistics: Student’s t-test, n = 2. (C) The indicated 4T1 spheroids and polypeptides were analyzed by IF-CM. In this and following figures denoted in arbitrary units (AU) are fluorescence intensities (FI) expressed as mean ± SD of five representative fields (n = 5). Statistics: Student’s t-test. (D) RQ of the indicated mRNA is represented as in B comparing A549-derived clone C79 (progenitor) or G7A (mesenchymal) expressing GPBP-EYFP with an A549 clone expressing EYFP (EYFP). Shown is a representative analysis of two independent experiments. Statistics: Student’s t-test, n = 2. In all the figures: n, sample size per group; *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001; DAPI (4′,6-diamidine-2′-phenylindole dihydrochloride) stained cell nucleus; WCIF ImageJ was used for quantification of WB; unless otherwise indicated scale bars were 50 μm.