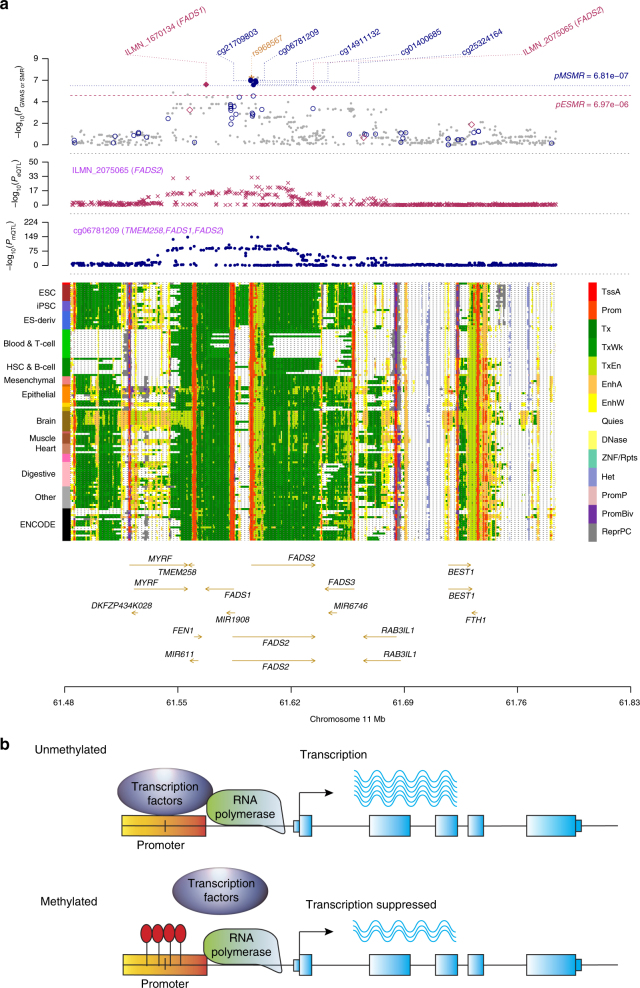
Fig. 4.
Prioritizing genes and regulatory elements at the FADS1/FADS2 locus for rheumatoid arthritis (RA) with a plausible regulation mechanism. a Results of SNP and SMR associations across mQTL, eQTL and GWAS. The top plot shows −log10(P-values) of SNPs from the GWAS meta-analysis for RA42. The red diamonds and blue circles represent −log10(P-values) from SMR tests for associations of gene expression and DNAm probes with RA, respectively. The solid diamonds and circles are the probes not rejected by the HEIDI test. The yellow star indicates the previously reported causal variant rs968567. The second plot shows −log10(P-values) of the SNP association for gene expression probe ILMN_2075065 (tagging FADS2) from the CAGE eQTL study. The third plot shows −log10(P-values) of the SNP associations for DNAm probe cg06781209 from the mQTL study. The bottom plot shows 14 chromatin state annotations (indicated by colours) of 127 samples from REMC for different primary cells and tissue types (rows). b A hypothetical regulation mechanism. When the DNAm site in the promoter is unmethylated, the transcription factor SREBF2 (activator) binds to the promoter and enhances the transcription of the FADS2 gene. When the DNAm site is methylated (by the effect of the genetic variant rs968567 at the promoter), the binding of SREBF2 is disrupted and therefore the transcription of FADS2 is suppressed