Fig. 2.
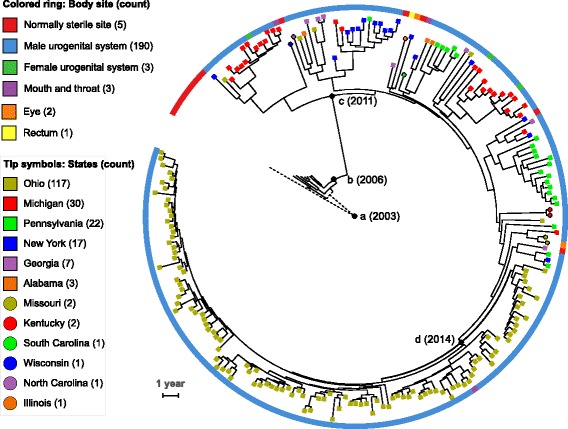
Time-measured Bayesian phylogeny U.S. NmNG urethritis clade. The analysis included 204 isolates belonging to the clade, with nine closely related invasive isolates used as an outgroup. Isolates are marked according to the state in which they were isolated (symbols on branch tips), and body site from which they were isolated (colored ring). Counts in the key refer to isolates in the U.S. NmNG urethritis clade. Nodes mentioned in the text are marked with black circles and labeled with the estimated year: (a) the root; (b) divergence between urethritis clade and NmC isolates, (c) the U.S. NmNG urethritis clade; and (d) the clade comprising solely isolates from Columbus, OH. The two branches originating at the root (node “a”) are dashed to indicate that inferred mutational events on the branch leading to the outgroup cannot be oriented with respect to time. The scale bar represents 1 year
