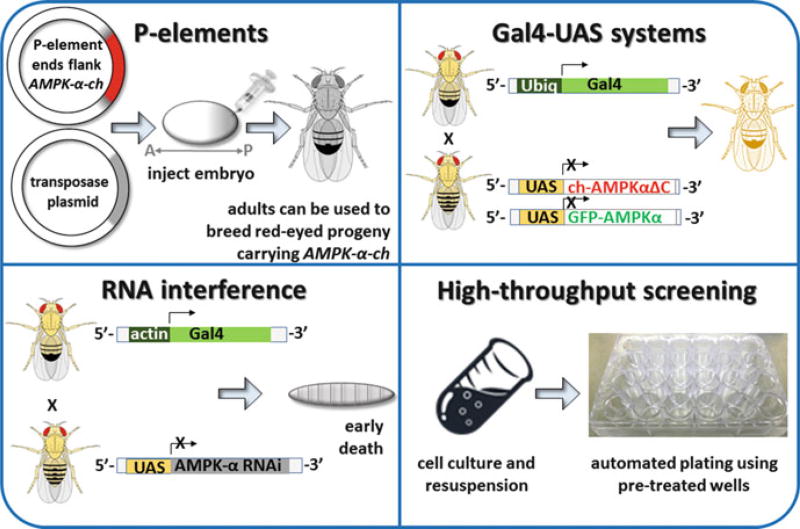
Fig. 16.3. Tools used in Drosophila research.
Techniques used by fly geneticists are illustrated. (Clockwise from top left) P-elements: A transgene plasmid (e.g., encoding cherry-tagged AMPK-α) and helper plasmid (encoding transposase) can be mixed and injected into Drosophila embryos (Bachmann and Knust 2008; Beall and Rio 1997). The inverted terminal repeats flanking the transgene are recognized by transposase, which randomly inserts the transgene into genomic DNA (Bachmann and Knust 2008; Beall and Rio 1997). Embryos will then develop into adults in which some germ cells carry a transgene insertion. Adult males can be crossed to females to generate offspring that retain the insertion in every cell (Bachmann and Knust 2008; Majumdar and Rio 2015). Drosophila offspring carrying the P-element insertion can be identified by a marker (typically a gene for red eyes) that is also encoded separately in the transgene plasmid (Bachmann and Knust 2008; Majumdar and Rio 2015). Gal4-UAS gene expression systems: Gal4 and UAS-transgene parental lines can be crossed to generate offspring in which transgene expression is driven by Gal4 (Brand and Perrimon 1993). Kazgan et al., for example, used Ubiq-Gal4 to drive ubiquitous (Ubiq) expression of both full-length GFP-tagged dAMPK-α and mCherry (ch)-tagged truncated dAMPK-α in offspring, thereby allowing them to identify a nuclear export signal in dAMPK-α (Kazgan et al. 2010). The figure depicting Kazgan et al.’ s technical approach is adapted from the earliest publication describing the Gal4-UAS system (Brand and Perrimon 1993). RNA interference: Gal4 can also be used to drive the expression of RNA (Perkins et al. 2015). In the example shown, knockdown of AMPK-α can be used to help identify changes in AMPK-dependent signaling pathways in larvae (Onyenwoke et al. 2012). High-throughput screening: Drosophila cell lines allow researchers to take advantage of Drosophila’s streamlined genome without having to breed stable fly lines. In the example shown, cells can be rapidly plated onto wells that have been pretreated with an RNAi library. Moser and colleagues have used this approach to screen for host factors (such as AMPK) that regulate viral infection (Moser et al. 2010a, b)