Abstract
The epidermal growth factor receptor (EGFR) is an important transmembrane glycoprotein kinase involved the initiation or perpetuation of signal transduction cascades within cells. These processes occur after EGFR binds to a ligand [epidermal growth factor (EGF)], thus inducing its dimerization and tyrosine autophosphorylation. Previous publications have highlighted the importance of glycosylation and dimerization for promoting proper function of the receptor and conformation in membranes; however, the effects of these associations on the protein conformational stability have not yet been described. Molecular dynamics simulations were performed to characterize the conformational preferences of the monomeric and dimeric forms of the EGFR extracellular domain upon binding to EGF in the presence and absence of N-glycan moieties. Structural stability analyses revealed that EGF provides the most conformational stability to EGFR, followed by glycosylation and dimerization, respectively. The findings also support that EGF–EGFR binding takes place through a large-scale induced-fitting mechanism.
Keywords: ErbB, glycoprotein, molecular dynamics, EGF binding
INTRODUCTION
The epidermal growth factor receptor (EGFR) family of transmembrane proteins, commonly known as ErbB receptors, is located on human mesothelial, lung epithelial cells and other epithelial cells throughout the body. This family of glycoproteins plays a critical role in initiation or perpetuation of signal transduction cascades within cells. An agonist that commonly binds to the extracellular domain of the EGFR is the epidermal growth factor (EGF), a small polypeptide hormone.1 After binding to EGF, the receptors dimerize to form either homodimers or heterodimers with other members of the ErbB family.2–4 This in turn activates the intracellular kinase domain of the EGFR, which after autophosphorylation, triggers a complex reaction of signals in the cytoplasm and nucleus. The physiological response to this signal transduction cascade is increased cell survival, proliferation and the inhibition of apoptosis,2 including the transactivation of proto-oncogenes important in mitogenesis.5 In addition, inhibition of the EGFR expression (e.g., patients under cancer therapy treatment) has been reported to promote skin infections by Staphylococcus aureus.6 The bacterium is also able to exploit the EGRF signaling pathway to evade immune responses,7 as well as to invade across epithelial barrier to disseminate itself into the bloodstream.8
The 170-kDa EGFR (or ErbB1) has an extracellular ligand-binding domain (~620 amino acids), a membrane-spanning region, and an intracellular cytoplasmic domain (~550 amino acids) that contains a tyrosine kinase domain.3 Along its extracellular region, 12 potential sites for N-glycosylation are reported.9 Several studies have shown that N-linked glycosylation of the ErbB proteins is an important factor in regulating their dimerization and function.10–15 In addition, previous molecular dynamics simulations of N-linked glycans have shown for a number of systems that glycosylation confers stability to the glycoprotein to which they are attached.16–18 Although complete processing of EGFR glycans to higher order mannose-type or complex-type oligosaccharides is not essential for protein function,19 the presence of simple glycans, such as mono- or disaccharides, were observed to help provide more stable configurations. Based on this observation, Takahashi et al. have postulated that N-glycans affect the conformation of ErbB family, which is crucial for their natural activity.14 In addition, unnatural overexpression of EGRF has been triggered by exposure to inorganic substances20–22 and associated with lung cancers and malignant mesotheliomas.20,21
Thus, considering the importance of the EGFR in both physiological processes and disease, classical molecular dynamics simulations were performed in order to better comprehend the structural variables that influence EGFR activity. For this purpose, the importance of N-glycosylation on the EGFR extracellular domain, as well as the effects of dimerization and EGF binding to the receptor was evaluated by means of dimeric and monomeric EGFR interaction with its natural ligand, in the presence and absence of N-linked oligosaccharides.
MATERIALS AND METHODS
Nomenclature, software, and analysis
All MD simulations of EGFR extracellular domain were performed using the GROMACS v.4.5.1 package.23 The GROMOS force field parameter set 53A6 in conjunction with its carbohydrate 45A4 extension24,25 was used for modeling the initial coordinates of the protein obtained from the Protein Data Bank (PDB ID code 1IVO),26 containing two copies of EGF hormone bound to a EGFR dimer, with residues ranging 2–512, thus lacking part of Domain IV and the entire transmembrane domain. The atom-positional root mean square displacement (RMSD) and the amino acid root mean square fluctuations (RMSF) were calculated considering the protein Cα atoms. The time-dependent secondary structure was calculated using the dictionary of secondary structure of proteins (DSSP) program,27 while all remaining analyses were performed using the postprocessing tools within the GROMACS package.23
N-glycosylation parameters and systems preparation
The EGFR dimer was prepared for molecular simulations by building topologic databases for its glycosylated asparagine residues. The heterogeneous residue N-acetyl-D-glucosamine was made readable by GROMACS by creating two new topologically distinct residues, ASN2 and ASN3, which has either one or two GlcNAc molecules attached to an Asn residue, respectively. The two sugar molecules in ASN3 have a beta 1 → 4 connection. The employed parameters were compiled based on previously validated GROMOS potential-energy and interaction terms, in which the monosaccharides were described as in GROMOS 45A4 parameters set,24 and the N-glycosydic linkage as in GROMOS 53A6glyc force field.17 Subsequently, the EGFR PDB ID 1IVO structure was modified to prepare the simulated systems to be a monomer or a dimer, glycosylated or nonglycosylated, and with or without its natural binding ligand (EGF). The composition and abbreviation for each system is shown in Table I. In the N-glycosylated systems, EGFR (Fig. 1) contained GlcNAc-linked residues as in PDB ID 1IVO.26 Therefore, the monomeric systems were glycosylated on the six possible N-glycosylation sites, located at asparagine residues 32, 151, 172, 328, 337, and 420. The dimeric systems were N-glycosylated on the six abovementioned sites on Monomer 1 and on three sites on Monomer 2. Sugars were added to asparagine glycosilation sites 32, 151, and 358 for the latter, as described in the crystal structure of EGFR PDB ID 1IVO. Although higher glycosylation states have not been used, it is known that 2/3 of the stabilization of the native state of a N-linked glycoprotein has been accounted for the first GlcNAc-linked monosaccharide, while the remaining 1/3 contribution to its structural stability comes from the second and third monosaccharides.28 Therefore, the use of mono- and disaccharide N-glycans is expected to account for their influence on EGFR stability
Table I.
Summary of the Simulated EGFR Systems
| System | Residues | Number of water molecules | Ionsa | Total number of atoms | Box volume (nm3) |
|---|---|---|---|---|---|
| dEGFR | 1020 | 131,232 | 6 | 403,822 | 4197 |
| dEGFRNAG | 1020 | 131,141 | 6 | 403,719 | 4197 |
| dEGFREGF | 1116 | 130,662 | 3 | 403,285 | 4197 |
| dEGFRNAG-EGF | 1116 | 130,754 | 3 | 403,809 | 4197 |
| mEGFRNAG | 510 | 55,064 | 3 | 170,374 | 1772 |
| mEGFRNAG-EGF | 558 | 54,804 | 3 | 170,101 | 1772 |
The ions present were Cl− in the ligand-free systems and Na+ in the ligand-bound EGFR.
Abbreviations: dEGFR, dimeric EGFR; dEGFRNAG, dimeric, glycosylated EGFR; dEGFREGF, dimeric, ligand-bound EGFR; dEGFRNAG-EGF, dimeric, glycosylated, ligand-bound EGFR; mEGFRNAG, monomeric, glycosylated EGFR; mEGFRNAG-EGF, monomeric, glycosylated, ligand-bound EGFR.
Figure 1.
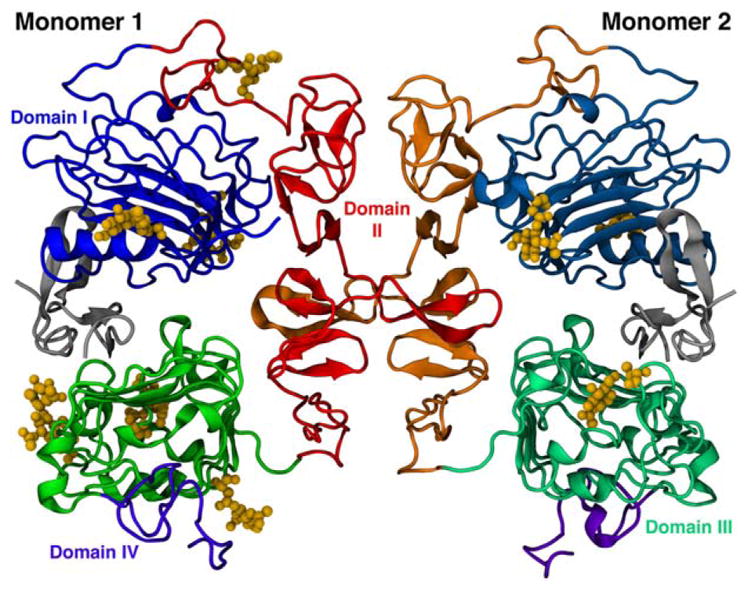
Ribbon representation of the dimeric form of EGFR extracellular domain. The arrangement of the four domains are represented by different shades of color in each monomer, while N-glycans are shown as yellow ball and sticks.
Molecular dynamics simulations
Each simulation system was placed in a box with periodic boundary conditions and solvated with explicit SPC water model molecules.29 The systems were neutralized with Na+ and Cl− ions when necessary (see Table I). The systems were initially energy optimized by 10,000 iterations of the steepest descent algorithm, followed by a 10-ns molecular dynamics equilibration time. A 50-ns production time was subsequently obtained for each system using a 2-fs time step integrated via the leapfrog algorithm.30 Simulations were done at constant pressure (1 atm) and temperature (310 K) using the Berendsen-barostat and thermostat, respectively.31 All bond lengths were constrained using the LINCS algorithm,32 whereas the geometry of the water molecules was constrained by the SETTLE algorithm.33 A 1.4-nm short-range cutoff was assigned to all van der Waals and electrostatic interactions and long-range electrostatic interactions were treated by the generalized reaction field method,34 using a εrf = 66. It is worth noting that is out of the scope of this work to fully characterize the EGFR dynamics. This has been previously done in a comprehensively manner.35–38 The simulation length was chosen based on observation of the phenomena studied, so that the analyses would leave no open questions into the subject of interest.
RESULTS AND DISCUSSION
EGFR conformational stability
For each of the six EGFR systems shown in Table I, we calculated an overall assessment of the protein structural behavior during MD simulations, the atom positional RMSD, the RMSF, and the secondary structure content and solvent accessible surface area (SASA), as assessment of these properties provide information on overall protein and binding site conformation and stability. As a general feature, neither N-glycosylation, dimerization or binding to EGF were observed to significantly influence the SASA or the secondary structure content of the EGFR in the performed simulations (Supporting Information Fig. S1 and Table SI). It indicates that these factors do not significantly influence intradomain EGFR stability.
Dimerization
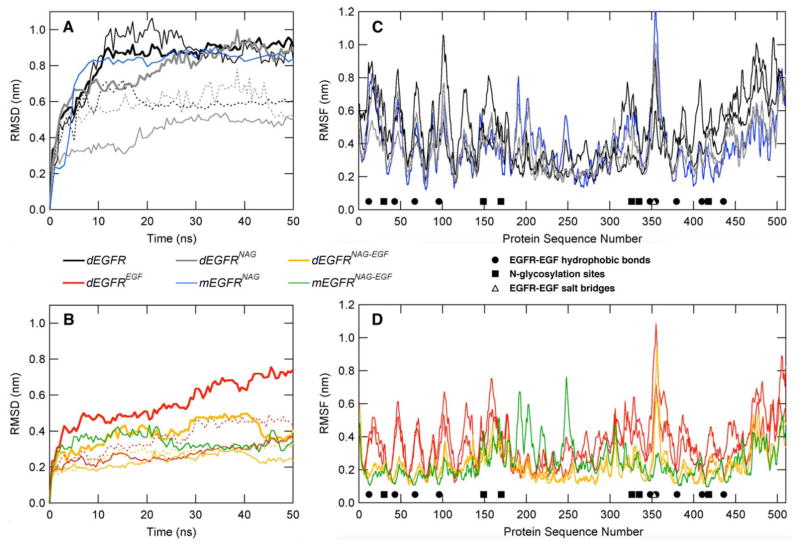
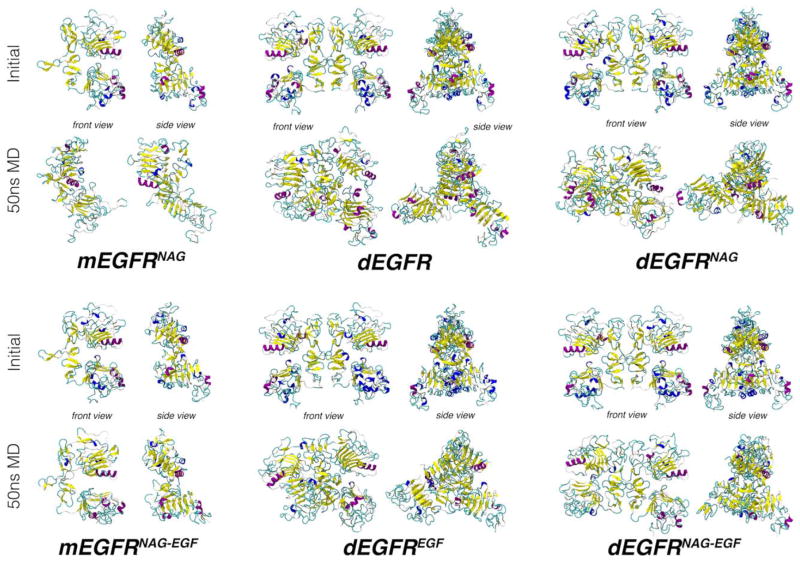
Regarding EGFR dimerization, a comparison of the two monomeric EGFR systems with the dimeric systems suggests a minimal effect of dimerization on the protein structure [Fig. 2(C) and (D)]. Physiologically, EGFR dimerization only occurs after ligand binding and, in such conditions, only small RMSD differences may be observed for the monomeric system [Fig. 2(B), green curve] and both monomers in the dimeric systems [Fig. 2(B), thin gold lines]. In the absence of EGF, on the other hand, both monomers in the dimeric system [Fig. 2(A), thin gray lines] showed significantly lower RMSDs than in the monomeric system [Fig. 2(A), blue curve]. In fact, the final structure of the mEGFRNAG system appears to have undergone much more conformational rearrangements then each monomer in the dEGFR system (Fig. 3).
Figure 2.
Conformational properties of the EGFR without (A and C) and with (B and D) the EGF ligand. In (A) and (B), root mean square deviations (RMSD) for EGFR αC as a function of time. Bold lines indicate RMSD calculations of dimeric EGFR, whereas thin solid lines show RMSD for Monomer 1 and the dashed lines show RMSD for Monomer 2, each calculated individually. In (C) and (D), per-residue root mean square fluctuation, in which both monomers are represented with the same line style for each monomer in dimeric systems. For (C) and (D), filled circles indicate hydrophobic interactions, open triangles show the positions of salt-bridges, and filled squares are N-glycan sites.
Figure 3.
Snapshots for the initial and final structures of the simulated EGFR are presented, in front and side views. In these structures, the EGF hormone, when present, was omitted for clarity. Proteins are represented in cartoon model, where α-helices are displayed in purple, helices3–10 in blue, β-sheets in yellow, loops in cyan, and unstructured regions in white.
Despite this, the RMSF analyses suggest that the flexibility of the monomeric systems are very similar in magnitude to the monomers in the dimeric systems [Fig. 2(C), blue and gray lines; Fig. 2(D), green and gold lines]. The exception is the region comprised by EGFR Domain II, from amino acids 166–309, related with the EGFR dimerization arm. This region showed an increased flexibility, especially around Regions 190–215 and Residue 250 [Fig. 2(C,D)]. In this case, the absence of the second monomer allowed the residues in this domain to show higher fluctuations.
N-Glycosylation
Recently, a complete monomeric EGFR glycosylated structure was studied through MD simulations embedded in a membrane bilayer.38 Such study identified that N-glycosylation is critical for the EGFR extracellular domain to properly interact with the membrane, and thus to maintain its adequate conformation.38 Our results corroborate to a global stabilization of the receptor due to glycosylation [Fig. 2(A,B)], especially when the receptor is bound to EGF. In addition, we observe that the removal of the glycans in the EGFR–EGF system yields a higher RMSD in the receptor dimer than in its glycosylated counterpart [Fig. 2(B), red and yellow bold lines].
Structurally, the absence of N-glycosylation disrupts the EGFR arrangement such that Domain III of the monomers shift away from each other (Fig. 3, dEGFREGF and dEGFRNAG-EGF). A similar rearrangement involving Domains III, but in a significantly smaller scale, has also been observed in another molecular dynamics simulations of a EGFR contained a modeled Domain IV, when in contact with a POPC membrane (but not in water).39 The authors find that overall Domain IV is highly flexible, while Domains I–III are highly stable. If the RMSD of dEGFREGF and dEGFRNAG-EGF is calculated for each monomer separately, both N-glycosylated polypeptides also tend to show lower RMSDs, especially in the final part of MD simulations [Fig. 2(B), straight and dashed, yellow thin lines] in comparison to their nonglycosylated counterparts [Fig. 2(B), straight and dashed, red thin lines]. Taken together, our results suggest N-glycosylation is crucial for Domain III stabilization.
When the protein is in the unbound state, the RMSD plots for the nonglycosylated and glycosylated dimeric proteins do not differ significantly [Fig. 2(A), black and gray bold curves]. However, in both systems the apparent increase in the distance between Domain III also occurs in the monomers (Fig. 3, dEGFR and dEGFRNAG). As suggested by the RMSD graphs, the presence of N-glycan seems to alter the flexibility of specific regions of the protein. Thus, local conformational stabilization effects are observed, in which the glycosylated receptor shows local lower flexibility profiles near N-glycosylation sites [Fig. 2(C,D), squares]. Furthermore, near the regions of hydrophobic binding to EGF, as around amino acids Leu12, Tyr43, Leu67, and Leu96, the systems where N-glycosylation is absent tend to be more flexible [Fig. 2(C), black versus gray curves; Fig. 2(D), red and gold curves]. These data indicate that N-glycosylation may also stabilize some of the residues involved in EGFR–EGF interaction.
EGFR–EGF binding
The EGFR extracellular domain may be divided into three main sections that represent its four structural subdomains (Fig. 1). Amino acids 1–165 make up domain and Section I, amino acids 166–309 compose domain and Section II, and the third section is composed of Domain III (amino acids 310–481) plus Domain IV (amino acids 482–510). The Sections I and III comprise the EGFR regions associated with EGF binding, whereas Section II, as previously commented, is associated with the dimerization arm used to homodimerize the EGFR or help it bind other ErbB receptors.
From the MD simulations, both Sections I and III display a peak-and-valley pattern, in which the peaks are hydrophobic residues (represented with filled circles) involved with EGF binding [Fig. 2(C,D)]. As expected, the highest fluctuations on these residues are observed in the systems lacking EGF [Fig. 2(C)]. Although the role of dimerization appear to be minimal, EGF binding seems to be crucial in the receptor stabilization, as visualized by the similarity of the starting structures (Fig. 3) and the marked difference in RMSD between the monomeric [Fig. 2(A), blue curve and Fig. 2(B), green curve] and the dimeric nonglycosylated [Fig. 2(A), black curve and Fig. 2(B), red curve] versus glycosylated [Fig. 2(A), gray curve and Fig. 2(B), gold curve] proteins.
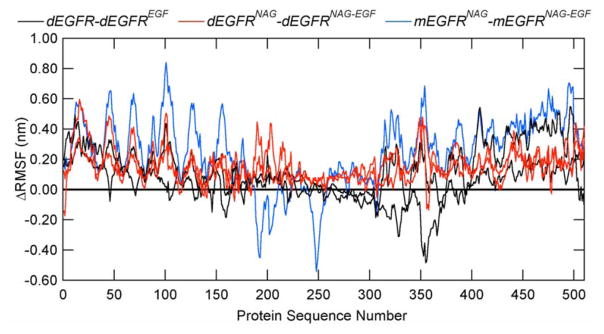
In terms of flexibility, while subtracting the RMSF values of EGF-absent and EGF-present systems, it may be observed that EGFR mobility is overall reduced due to the presence of its natural ligand (Fig. 4), as most of the resultant ΔRMSF graphs contain positive values. Besides minor regions in Domain I (Residues 1–165), two worth noting exceptions are observed: (1) the regions in the dimerization arm in Domain II in the monomeric systems (Fig. 4, blue line, between Residues 180 and 270), possibly due to the lack of the complementary monomer in these systems and (2) Domain III in the dimeric and glycan-free systems (Fig. 4, black curve, between Residues 300 and 400), in which the N-glycosylation sites lie three-dimensionally close to the EGF-binding pocket (Fig. 1). The latter results add to the previously discussed data on the importance of N-glycosylation in proper receptor–ligand interaction, especially regarding the stabilization of the region around Residue 350 on EGFR Domain III in the presence of the EGF ligand, where two residues involved in EGFR–EGF hydrophobic bonds and one in EGFR–EGF salt bridge are located.
Figure 4.
The difference in RMSF of EGFR upon binding to the ligands shown for dEGFR minus dEGFREGF (black curve), dEGFRNAG minus dEGFRNAG-EGF (red curve), and mEGFRNAG minus mEGFRNAG-EGF (blue curve). Zero line in bold black is shown for reference. A positive value indicates a decrease in RMSF upon addition of the ligand to the simulated EGFR. Monomers 1 and 2 are shown by the same line style for the dimeric EGFR systems.
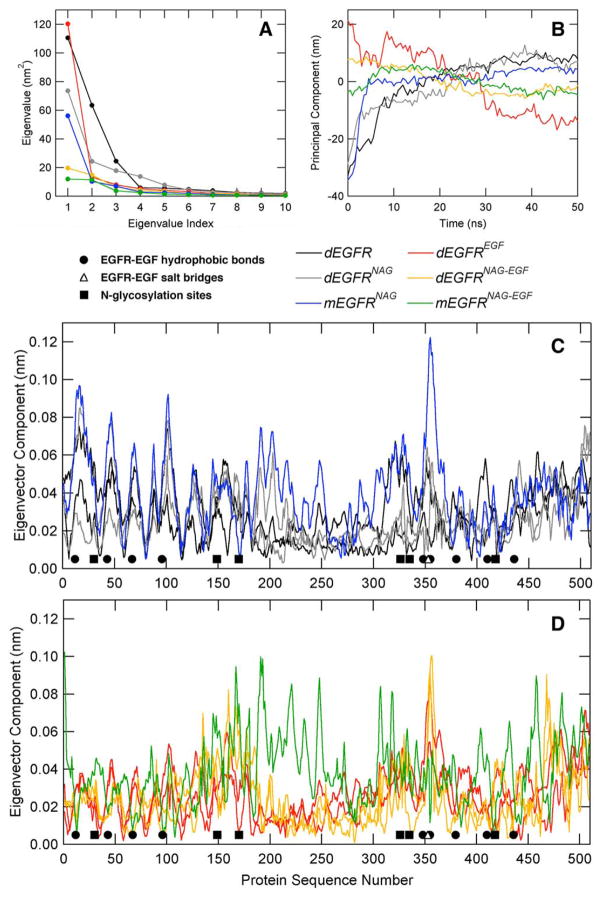
In order to further explore the EGFR conformational dynamics, a principal component analysis (PCA) was performed. As a result, most of the identified amplitude motions are mostly represented by the first 3–4 modes [Fig. 5(A)]. Additionally, the systems containing both N-glycans and EGF show the lowest first eigenvalue [Fig. 5(A), green and gold lines], followed by the N-glycosylated systems [Fig. 5(A), blue and gray lines]. The principal components of EGFR Cα indicate that the systems devoid of EGF rise in value as the simulation progresses [Fig. 5(B), blue, black, and gray lines], whereas EGF-bound systems show a steady decline or maintenance [Fig. 5(B), red, green, and gold lines]. This finding is in agreement with the fact that EGFR in an unbound state is largely monomeric and with the mechanistic model that binding of EGF is associated with a dramatic conformational change.39,40 At the residue-level, projecting the first eigenvectors onto the EGFR sequence, the parts of the protein most involved in large-amplitude motions become more evident [Fig. 5(C,D)]. Therefore, as observed from the RMSF plots [Fig. 2(C,D)], high fluctuations are observed in the dimerization arm, located in Domain II, for the monomeric systems [Fig. 5(C,D)], while a relatively noisy behavior is observed for the regions embracing Domains III and IV. Nevertheless, the main difference is related to the N-terminal segment of Domain I, residues about 1–100, which present high flexibility in the absence of EGF and reduced motions in the presence of EGF [Fig. 5(C,D)], including amino acids involved in the EGFR:EGF binding, as residues Leu12, Tyr43, Leu67, and Leu96.
Figure 5.
Principal component analysis (PCA) for the performed simulations. (A) EGFR eigenvalues for the most representative eigenvectors, as calculated from the coordinates of the α-carbon atoms. (B) Projection over α-carbon atom trajectories. (C) and (D) EGFR eigenvector projection over protein sequence. For the dimeric proteins, both monomers are represented with the same line style.
EGF binding pocket
The space between Domains I and III of EGFR extracellular domain comprises the binding pocket to EGF and other analogous ligands (Fig. 6). In order to further evaluate the influence of dimerization, N-glycosylation and EGF-presence, the minimum distance between residues L12 and V348 was measured, as a metric for binding pocket width, throughout the simulations (Fig. 6). In agreement with the previously discussed results, the strongest effects on the interdomain distance are caused by the presence of EGF, causing the space between L12 and V348 to be smaller in such systems (Fig. 6, green, orange, and red bars). Such data indicate that EGFR–EGF interaction hinders the hydrophobic residues from solvent and tighten the opening between domains, while EGF removal disrupts the binding pocket (Fig. 6, black, gray, and blue bars).
Figure 6.
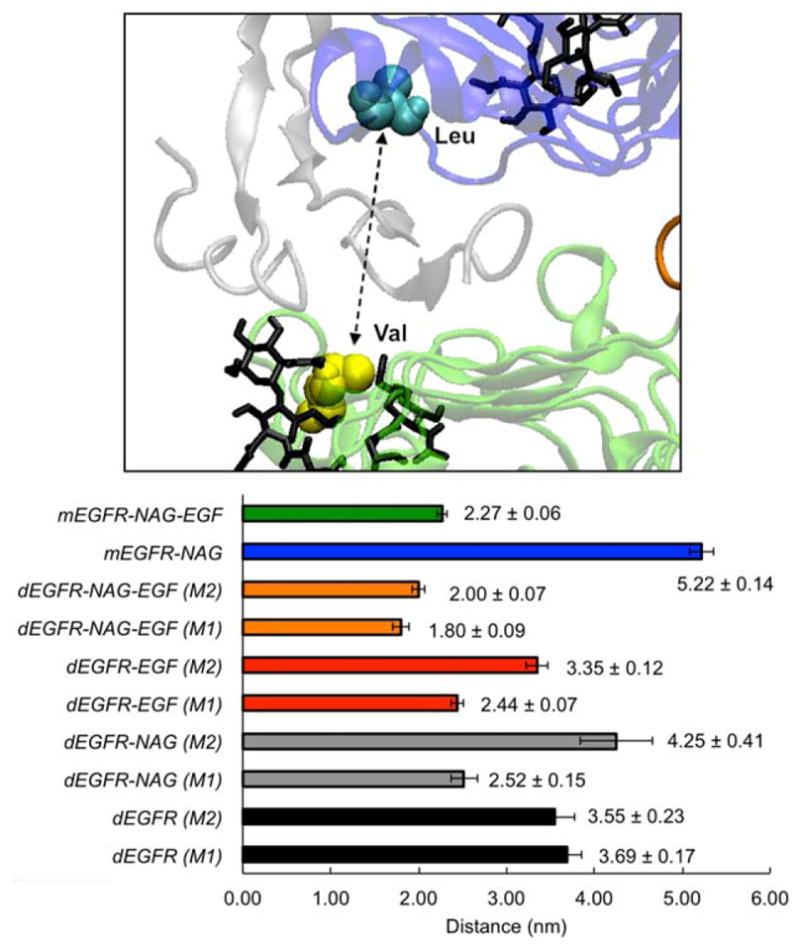
EGFR–EGF binding pocket behavior during molecular dynamics simulations. Upper figure depicts the EGF binding pocket, flanked by Residues L12 and V348. The lower graph shows the average distances (along with its standard deviation) between EGFR residues L12 and V348 for all systems between the 30 and 50 ns time.
Furthermore, N-glycosylation appears to contribute in the maintenance of a better binding pocket organization for EGFR–EGF binding, as the N-glycosylated receptor has lower distance values than nonglycosylated counterparts (Fig. 6, red versus orange bars, and green versus blue bars). On the other hand, such difference is not so apparent when EGF is absent, as both higher and lower distance values are observed in the glycosylated monomers (Fig. 6, gray bars) than in the nonglycosylated ones (Fig. 6, black bars).
CONCLUSIONS
The transduction of extracellular signals by the cell surface receptor EGFR is an important factor in the initiation or perpetuation of signal transduction cascades within cells. Such signals regulate natural processes such as cell growth and development. They may also be associated with oncogenesis (e.g., mesothelioma) through aberrant activation of the cell signaling cascades. A part of this signaling cascade is the activation of EGFR through binding of either biological compounds, such as extracellular ligands (i.e., EGF), or foreign substances.41 Upon binding to a ligand, the receptor homo- or heterodimerizes and autophosphorylation start the signal to the nucleus. The complexity of EGFR dynamics upon binding of different ligands has been recently addressed and a similar binding mode was observed.42 Therefore, the aim of this study was to assess structural variables (dimerization, N-glycosylation and substrate binding) that influence EGFR structural stability and the importance of N-glycosylation on the protein’s extracellular domain.
Several experimental studies have reported the importance of N-glycosylation on the functional properties of EGFR, including cell surface expression,12,13 ligand binding,12,13,43,44 dimerization,14,15,45–47 interaction with membranes,38 and endocytosis.44,48 These properties are intrinsically related to EGFR conformation, such that an inadequate protein conformation may impair the receptor to perform such activities. However, the degree of conformational change linked to EGFR function has not been inferred. The present study is the first atomic-level report of a comparison between nonglycosylated and glycosylated EGFR molecules, both in monomeric and dimeric forms.
Analyses of the simulations showed that all three variables played a decisive role in the conformational stability of the EGFR within the simulation time, highlighting their relative importance on EGFR stability. The effect of dimerization yielded the least amount of change in tertiary structure and conformational dynamics. Protein N-glycosylation showed a more significant impact, especially when the protein was in its dimeric form. This finding confirms the importance of N-linked glycans in stabilizing protein conformation,16–18 and aligns with the mechanistic model that binding of EGF is associated with a dramatic conformational change.39,40
Altogether, the influence of the three variables in this study to EGFR extracellular domain conformational stabilization seems to be hierarchal, whereby the presence of a ligand provides the major conformational stability to the receptor, followed by N-glycosylation, and lastly the dimerization of the protein. Such results are thus expected to provide an important reference for further molecular investigation into the EGFR dynamics upon exposure to other natural and foreign ligands.
Supplementary Material
Acknowledgments
LPF and RDL acknowledge support from CNPq, CAPES, and FACEPE (including grant number APQ-0398–1.06/13). SKL gratefully acknowledges financial support from the National Institutes of Health (R01HL119648) and the National Science Foundation (EAR0745808 and 1424138). Partial computer allocation was granted by the Environmental Molecular Sciences Laboratory (EMSL), a US Department of Energy facility housed at the Pacific Northwest National Laboratory (PNNL), and the Brazilian National Scientific Computing Center (LNCC).
Footnotes
This work was performed at The Ohio State University
Additional Supporting Information may be found in the online version of this article.
References
- 1.Wilkinson JC, Stein RA, Guyer CA, Beechem JM, Stros JV. Real-time kinetics of ligand/cell surface receptor interactions in living cells: binding of epidermal growth factor to the epidermal growth factor receptor. Biochemistry. 2001;40:10230–10242. doi: 10.1021/bi010705t. [DOI] [PubMed] [Google Scholar]
- 2.Ciardiello F, Tortora G. Drug therapy: EGFR antagonists in cancer treatment. New Engl J Med. 2008;358:1160–1174. doi: 10.1056/NEJMra0707704. [DOI] [PubMed] [Google Scholar]
- 3.Lemmon MA. Ligand-induced ErbB receptor dimerization. Exp Cell Res. 2009;315:638–648. doi: 10.1016/j.yexcr.2008.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schmitz KR, Ferguson KM. Interaction of antibodies with ErbB receptor extracellular regions. Exp Cell Res. 2009;315:659–670. doi: 10.1016/j.yexcr.2008.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cobb MH, Goldsmith EJ. How MAP kinases are regulated. J Biol Chem. 1995;270:14843–14846. doi: 10.1074/jbc.270.25.14843. [DOI] [PubMed] [Google Scholar]
- 6.Simanski M, Rademacher F, Schroder L, Glaser R, Harder J. The inflammasome and the epidermal growth factor receptor (EGFR) are involved in the Staphylococcus aureus-mediated induction of IL-1alpha and IL-1beta in human keratinocytes. PLoS ONE. 2016;11:e0147118. doi: 10.1371/journal.pone.0147118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gómez MI, Seaghdha MO, Prince AS. Staphylococcus aureus protein A activates TACE through EGFR-dependent signaling. EMBO J. 2007;26:701–709. doi: 10.1038/sj.emboj.7601554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Soong G, Martin FJ, Chun J, Cohen TS, Ahn DS, Prince A. Staphylococcus aureus protein A mediates invasion across airway epithelial cells through activation of RhoA GTPase signaling and proteolytic activity. J Biol Chem. 2011;286:35891–35898. doi: 10.1074/jbc.M111.295386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smith KD, Davies MJ, Bailey D, Renouf DV, Hounsell EF. Analysis of the glycosylation patterns of extracellular domain of the epidermal growth factor receptor expressed in Chinese Hamster ovary fibroblasts. Growth Factors. 1996;13:121–132. doi: 10.3109/08977199609034572. [DOI] [PubMed] [Google Scholar]
- 10.Matsumoto K, Yokote H, Arao T, Maegawa M, Tanaka K, Fujita Y, Shimizu C, Hanafusa T, Fujiwara Y, Nishio L. N-glycan fucosylation of epidermal growth factor receptor modulated receptor activity and sensitivity to epidermal growth factor receptor tyrosine kinase inhibitor. Cancer Sci. 2008;99:1611–1617. doi: 10.1111/j.1349-7006.2008.00847.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schlessinger J, Hakomori S. Ganglioside-mediated modulation of cell growth—specific effects of GM3 on tyrosine phosphorylation of the epidermal growth factor receptor. J Biol Chem. 1986;261:2434–2440. [PubMed] [Google Scholar]
- 12.Slieker LJ, Martensen TM, Lane MD. Synthesis of epidermal growth factor receptor in human A431-cells—glycosylation-dependent acquisition of ligand-binding activity occurs posttranslationally in the endoplasmic reticulum. J Biol Chem. 1986;261:5233–5241. [PubMed] [Google Scholar]
- 13.Soderquist AM, Carpenter G. Glycosylation of the epidermal-growth factor receptor in A-431 cells. The contribution of carbohydrate receptor function. J Biol Chem. 1984;259:2586–2594. [PubMed] [Google Scholar]
- 14.Takahashi M, Yokow S, Asahi M, Lee SH, Li W, Osumi D, Miyoshi E, Taniguchi N. N-glycan of ErbB family plays a crucial role in dimer formation and tumor promotion. BBA-Gen Subjects. 2008;1780:520–524. doi: 10.1016/j.bbagen.2007.10.019. [DOI] [PubMed] [Google Scholar]
- 15.Tsuda T, Ikeda Y, Taniguchi N. The Asn-420-linked sugar chain in human epidermal growth factor receptor suppresses ligand-independent spontaneous oligomerization—possible role of a specific chain in controllable receptor activation. J Biol Chem. 2000;275:21988–21994. doi: 10.1074/jbc.M003400200. [DOI] [PubMed] [Google Scholar]
- 16.Pol-Fachin L, Verli H. Assessment of glycoproteins dynamics from computer simulations. Mini-Rev Org Chem. 2011;8:229–238. [Google Scholar]
- 17.Pol-Fachin L, Verli H, Lins RD. Extension and validation of the GROMOS 53A6(GLYC) parameter set for glycoproteins. J Comput Chem. 2014;35:2087–2095. doi: 10.1002/jcc.23721. [DOI] [PubMed] [Google Scholar]
- 18.Lee HS, Qi Y, Im W. Extension and validation of the GROMOS 53A6(GLYC) parameter set for glycoproteins. Scientific Reports. 2015;5:8926. doi: 10.1002/jcc.23721. [DOI] [PubMed] [Google Scholar]
- 19.Stroop C, Weber W, Nimtz M, Gallego R, Kamerling J, Vliegenthart J. Fucosylated hybrid-type N-glycans on the secreted human epidermal growth factor receptor from swainsonine-treated A431 cells. Arch Biochem Biophys. 2000;374:42–51. doi: 10.1006/abbi.1999.1660. [DOI] [PubMed] [Google Scholar]
- 20.Faux SP, Houghton CE. Cell Signaling in Mesothelial Cells by Asbestos: Evidence for the Involvement of Oxidative Stress in the Regulation of the Epidermal Growth Factor Receptor. Inhal Toxicol. 2000;12:327–336. doi: 10.1080/08958378.2000.11463242. [DOI] [PubMed] [Google Scholar]
- 21.Faux SP, Houghton CE, Hubbard A, Patrick G. Increased expression of epidermal growth factor receptor in rat pleural mesothelial cells correlates with carcinogenicity of mineral fibres. Carcinogenesis. 2000;21:2275–2280. doi: 10.1093/carcin/21.12.2275. [DOI] [PubMed] [Google Scholar]
- 22.Zanella CL, Posada J, Tritton TR, Mossman BT. Asbestos causes stimulation of the extracellular signal-regulated kinase 1 mitogen-activated protein kinase cascade after phosphorylation of the epidermal growth factor receptor. Cancer Res. 1996;56:5334–5338. [PubMed] [Google Scholar]
- 23.Hess B, Kutzner C, van der Spoel D, Lindahl E. GROMACS 4: algorithm for highly efficient, load-balanced, and scalable molecular simulation. J Chem Theory Comput. 2008;4:435–447. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- 24.Lins RD, Hunenberger PH. A new GROMOS force field for hexopyranose-based carbohydrates. J Comput Chem. 2005;26:1400–1412. doi: 10.1002/jcc.20275. [DOI] [PubMed] [Google Scholar]
- 25.Oostenbrink C, Soares TA, van der Vegt NFA, van Gunsteren WF. Validation of the 53A6 GROMOS force field. Eur Biophys J. 2005;34:273–284. doi: 10.1007/s00249-004-0448-6. [DOI] [PubMed] [Google Scholar]
- 26.Ogiso H, Ishitani R, Nureki O, Fukai S, Yamanaka M, Kim JH, Saito K, Sakamoto A, Inoue M, Shirouzu M, Yokoyama S. Crystal structure of the complex human epidermal growth factor and receptor extracellular domains. Cell. 2002;110:775–787. doi: 10.1016/s0092-8674(02)00963-7. [DOI] [PubMed] [Google Scholar]
- 27.Kabsch W, Sander C. Dictionary of protein secondary structure—pattern-recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 28.Hanson SR, Culyba EK, Hsu T-L, Wong C-H, Kelly JW, Powers ET. The core trisaccharide of an N-linked glycoprotein intrinsically accelerates folding and enhances stability. Proc Nat Acad Sci USA. 2009;81:3684–3690. doi: 10.1073/pnas.0810318105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Berendsen HJC, Grigera JR, Straatsma TP. The missing term in effective pair potentials. J Phys Chem. 1987;91:6269–6271. [Google Scholar]
- 30.van Gunsteren WF, Berendsen HJC. A Leap-frog Algorithm for Stochastic Dynamics. Mol Simul. 1988;1:173–185. [Google Scholar]
- 31.Berendsen HJC, Postma JPM, van Gunsteren WF, Dinola A, Haak JR. Molecular dynamics with coupling to an external bath. J Chem Phys. 1984;81:3684–3690. [Google Scholar]
- 32.Hess B, Bekker H, Berendsen HJC, Fraaije J. LINCS: a linear constraint solver for molecular simulations. J Comput Chem. 1997;18:1463–1472. [Google Scholar]
- 33.Miyamoto S, Kollman PA. SETTLE—an analytical version of the SHAKE and RATTLE algorithm for rigid water models. J Comput Chem. 1992;13:952–962. [Google Scholar]
- 34.Tironi IG, Sperb R, Smith PE, van Gunsteren WF. A generalized reaction field method for molecular dynamics simulations. J Chem Phys. 1995;102:5451–5459. [Google Scholar]
- 35.Arkhipov A, Shan Y, Das R, Endres NF, Eastwood MP, Wemmer DE, Kuriyan J, Shaw D. Architecture and membrane interactions of the EGF receptor. Cell. 2013;152:557–569. doi: 10.1016/j.cell.2012.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Arkhipov A, Shan Y, Kim ET, Shaw D. Membrane interaction of bound ligands contributes to the negative binding cooperativity of the EGF receptor. PLos Comput Biol. 2014;10:e1003742. doi: 10.1371/journal.pcbi.1003742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Endres NF, Das R, Smith AW, Arkhipov A, Kovacs E, Huang Y, Pelton JG, Shan Y, Shaw DE, Wemmer DE, Groves JT, Kuriyan J. Conformational coupling across the plasma membrane in activation of the EGF receptor. Cell. 2013;152:543–556. doi: 10.1016/j.cell.2012.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kaszuba K, Grzybek M, Orlowski A, Danne R, Róg T, Simons K, Coskun U, Vattulainen I. N-glycosylation as determinant of epidermal growth factor receptor conformation in membranes. Proc Nat Acad Sci USA. 2015;112:4334–4339. doi: 10.1073/pnas.1503262112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kastner J, Loeffler HH, Roberts SK, Martin-Fernandez ML, Winn MD. Ectodomain orientation, conformational plasticity and oligomerization of ErbB1 receptors investigated by molecular dynamics. J Struct Biol. 2009;167:117–128. doi: 10.1016/j.jsb.2009.04.007. [DOI] [PubMed] [Google Scholar]
- 40.Lemmon MA, Schlessinger J, Ferguson KM. The EGFR family: not so prototypical receptor tyrosine kinases. Cold Spring Harb Perspect Biol. 2015;6:a020768. doi: 10.1101/cshperspect.a020768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Taylor ES, Wylie AG, Mossman BT, Lower SK. Repetitive dissociation from crocidolite asbestos acts as persistent signal for epidermal growth factor receptor. Langmuir. 2013;29:6323–6330. doi: 10.1021/la400561t. [DOI] [PubMed] [Google Scholar]
- 42.Sanders JM, Wampole ME, Thakur ML, Wickstrom E. Molecular Determinants of Epidermal Growth Factor Binding: A Molecular Dynamics Study. PLos One. 2013;8:e54136. doi: 10.1371/journal.pone.0054136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang X, Sun P, O’Gorman M, Tai T, Paller A. Epidermal growth factor receptor glycosylation is required for ganglioside GM3 binding and GM3-mediated suppression of activation. Glycobiology. 2001;11:515–522. doi: 10.1093/glycob/11.7.515. [DOI] [PubMed] [Google Scholar]
- 44.Sato Y, Takahashi M, Shibukawa Y, Jain SK, Hamaoka R, Miyagawa J-I, Yaginuma Y, Honke K, Ishikawa M, Taniguchi N. Overexpression of N-acetylglucosaminyltransferase III enhances the epidermal growth factor-induced phosphorylation of ERK in HeLaS3 cells by up-regulation of the internalization rate of the receptors. J Biol Chem. 2001;276:11956–11962. doi: 10.1074/jbc.M008551200. [DOI] [PubMed] [Google Scholar]
- 45.Yokoe S, Takahashi M, Asahi M, Lee SH, Li W, Osumi D, Miyoshi E, Taniguchi N. The Asn418-linked N-glycan of ErbB3 plays a crucial role in preventing spontaneous heterodimerization and tumor promotion. Cancer Res. 2007;67:1935–1942. doi: 10.1158/0008-5472.CAN-06-3023. [DOI] [PubMed] [Google Scholar]
- 46.Fernandes H, Cohen S, Bishayee S. Glycosylation-induced conformational modification positively regulates receptor-receptor association: a study with an aberrant epidermal growth factor receptor (EGFRvIII/DeltaEGFR) expressed in cancer cells. J Biol Chem. 2001;276:5375–5383. doi: 10.1074/jbc.M005599200. [DOI] [PubMed] [Google Scholar]
- 47.Whitson KB, Whitson SR, Red-Brewer ML, McCoy AJ, Vitali AA, Walker F, Johns TG, Beth AH, Staros JV. Functional effects of glycosylation at Asn-579 of the epidermal growth factor receptor. Biochemistry. 2005;44:14920–14931. doi: 10.1021/bi050751j. [DOI] [PubMed] [Google Scholar]
- 48.Lau KS, Partridge EA, Grigorian A, Silvescu CI, Reinhold VN, Demetriou M, Dennis JW. Complex N-glycan number and degree of branching cooperate to regulate cell proliferation and differentiation. Cell. 2007;129:123–134. doi: 10.1016/j.cell.2007.01.049. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.