Figure 1. Using CRISPR/Cas9 to stably integrate the HIV-NanoLuc provirus at the ROSA26 locus.
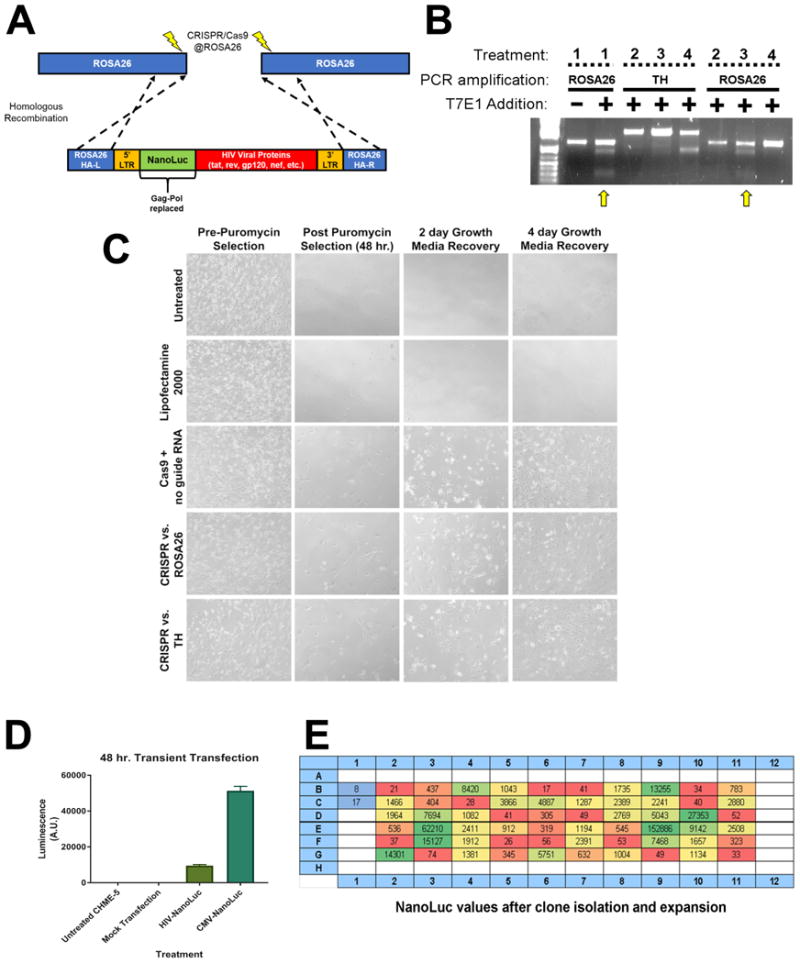
(A) Model of modified HIV provirus targeting to ROSA26 locus. Cas9 nickase is used to create mutagenic events at ROSA26. The HIV-NanoLuc provirus is flanked by ROSA26 homologous arms to facilitate integration. (B) CHME5 cells were transfected with CRISPR/Cas9 components and gRNAs targeting the ROSA26 locus, or tyrosine hydroxylase (TH) as a control. Puromycin selection was performed to select cell populations that were successfully transfected. PCR was then performed on genomic DNA to amplify the ROSA26 or TH genomic sequences. T7 endonuclease 1 enzyme was added to identify mutagenic events, and shows specific targeting of the ROSA26 locus in CHME-5 microglia by CRISPR/Cas9. Treatments are as follows 1: PC12 cells + ROSA26 gRNA (positive control), 2: CHME-5 cells + no gRNA, 3: CHME-5 cells + ROSA26 gRNA, 4: CHME-5 cells + TH gRNA. (C) Photomicrographs of CHME-5 cells undergoing puromycin selection. (D) Transient transfection of the HIV-NanoLuc provirus in CHME-5 microglia results in luciferase activity. Mock transfection of pBSII or a CMV driven NanoLuc were used as a negative and positive control respectively. (E) CRISPR/Cas9 at the ROSA26 locus was performed with the addition of the donor HIV-nanoLuc provirus. After puromycin selection, cells were clonally isolated in a 96 well plate and assayed for luciferase activity. Wells B1, B2: Parental CHME-5, Red wells: low NanoLuc expression, Yellow wells: medium NanoLuc expression, Green wells: High NanoLuc expression.
