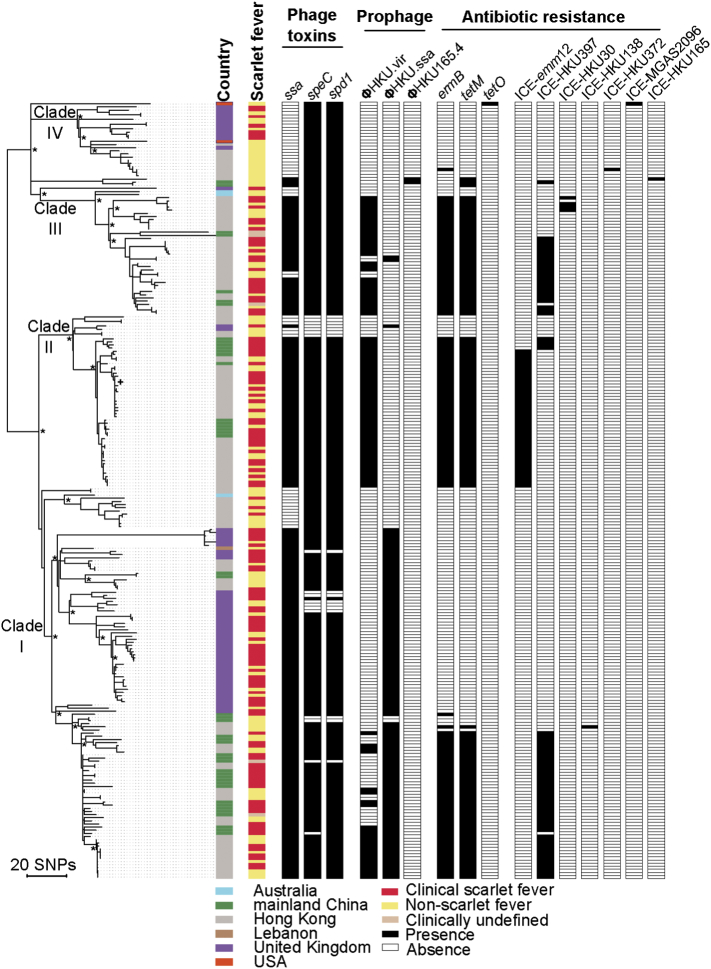
Fig. 5.
Midpoint-rooted maximum-likelihood phylogenetic tree of 248 GAS emm12 isolates based on 2637 polymorphisms after mapping to the Hong Kong scarlet fever reference genome HKU163 and removal of confounding genomic regions (see methods). Phylogenetic location of the HKU16 genome is represented by the cross. Previously defined phylogenetic clades of the Hong Kong emm12 lineage, termed Clades I-IV, are shown on major branches (Davies et al., 2015). Asterisks represent major nodes with > 95% bootstrap support. Terminal nodes are coloured by country of origin and clinical scarlet fever association. Relative distribution of selected virulence genes, prophage elements, antimicrobial resistance genes and integrative conjugative elements (ICE) that have previously been linked to the emergence of scarlet fever clones in Hong Kong(Davies et al., 2015) are displayed.