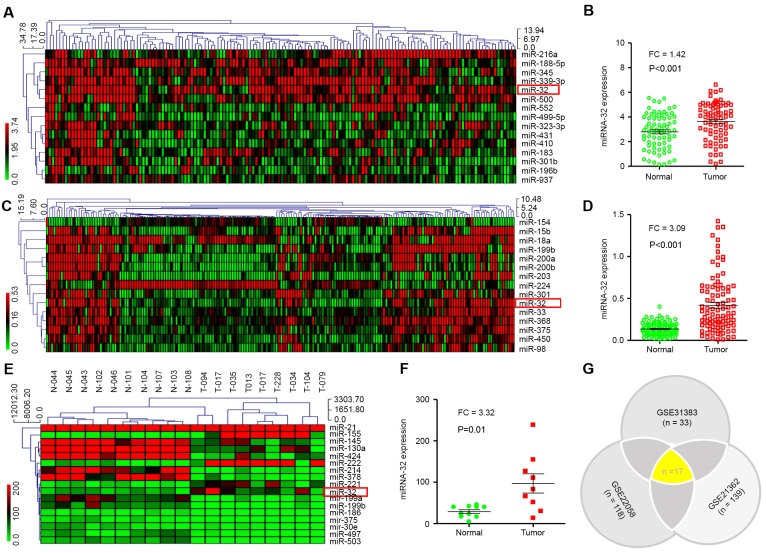
Figure 1.
Analysis of miR-32 expression in patients with HCC was performed using GEO datasets, and MEV4.7.1 clustering software was used to analyze different miRNAs. (A) Clustering analysis was performed using the MEV4.7.1 based on 15 dysregulated miRNAs using FC ≥1.4 and P<0.01. (B) miR-32 expression levels in HCC tissues vs. normal tissues were analyzed using the GEO database (GEO accession no. GSE21362). (C) Clustering analysis was performed with the MEV4.7.1 software. A total of 15 dysregulated miRNAs were screened using FC ≥3 or ≤0.3 and P<0.01. (D) The expression levels of miR-32 in HCC (n=96) and normal tissues (n=96) derived from the GEO database (GEO accession no. GSE22058) were analyzed. (E) Clustering analysis was performed using MEV4.7.1 based on 17 dysregulated miRNAs and screened using FC ≥3 or ≤0.3 and P<0.01. (F) miR-32 expression in HCC tissues (n=9) was significantly higher compared with non-tumor tissues (n=10). Datasets derived from the GEO database (GEO accession no. GSE22058). (G) A Venn diagram was generated using three GEO datasets. A total of 17 common miRNAs were identified, including miR-32. HCC, hepatocellular carcinoma; miR, microRNA; FC, fold-change; GEO, Gene Expression Omnibus; miRNA, microRNA.