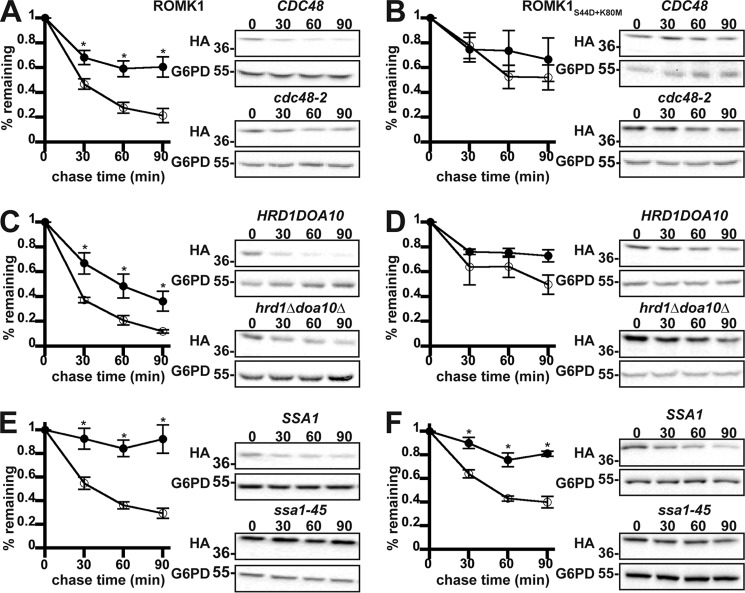
Figure 3.
ROMK1 is degraded by the ERAD pathway to a greater extent than ROMK1S44D+K80M in yeast. Yeast cultures expressing HA-tagged versions of either ROMK1 (A, C, and E) or ROMK1S44D+K80M (B, D, and F) grown to mid-logarithmic phase were dosed with cycloheximide, and aliquots were withdrawn at 0, 30, 60, and 90 min. Yeast strains containing either a temperature-sensitive mutation in the gene encoding Cdc48 (cdc48-2, filled circles) or lacking the mutant allele (CDC48, open circles) expressing wildtype ROMK1 (A) or ROMK1S44D+K80M (B) were shifted from room temperature to 39 °C for 60 min before addition of cycloheximide. A yeast strain lacking the ER-resident E3 ubiquitin ligases (hrd1Δdoa10Δ, filled circles) or an isogenic wildtype strain (HRD1DOA10, open circles) expressing wildtype ROMK1 (C) or ROMK1S44D+K80M (D) were shifted from room temperature to 37 °C 30 min before addition of cycloheximide. Yeast strains lacking four cytosolic isoforms of Hsp70 (Ssa1, Ssa2, Ssa3, and Ssa4) and containing either a temperature-sensitive mutation of Ssa1 (ssa1-45, filled circles) or an isogenic wildtype allele (SSA1, open circles) and expressing wildtype ROMK1 (E) or ROMK1S44D+K80M (F) were shifted from room temperature to 37 °C 30 min before addition of cycloheximide. For all experiments, ROMK expression was assessed by Western blotting and normalized to the initial time point. Representative images are shown, and all blots were stripped and reprobed for glucose-6-phosphate dehydrogenase (G6PD) as a loading control. Data represent the means of six independent experiments from two independent yeast transformations. Error bars show standard errors of the mean, and *indicates a significant (p < 0.05) difference as assessed by Student's t test.