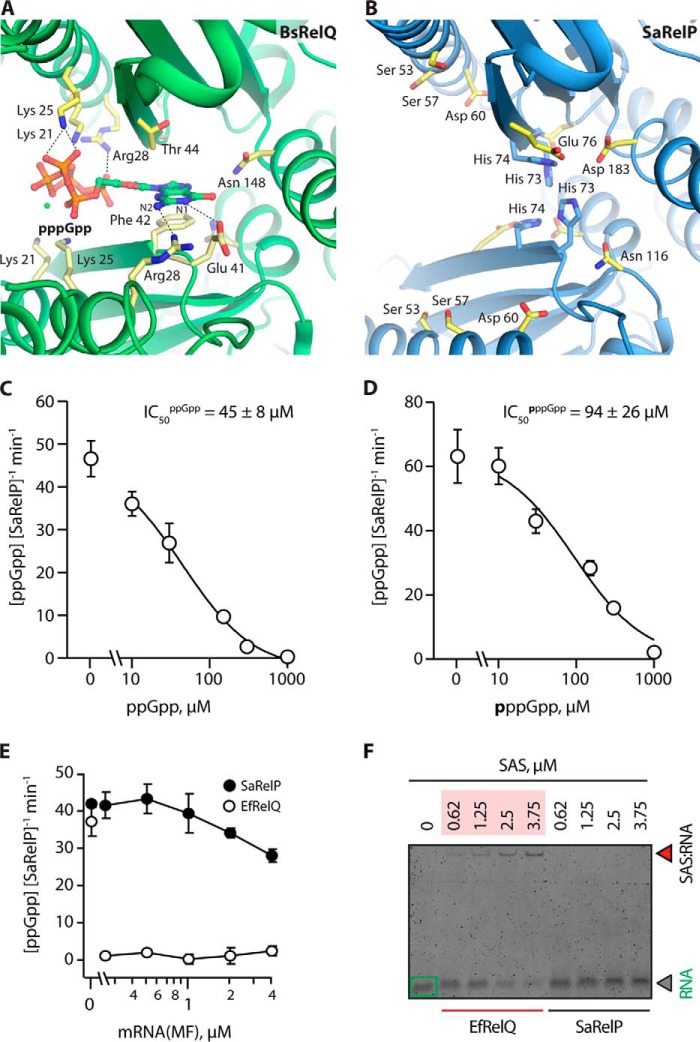
Figure 3.
SaRelP is negatively regulated by (p)ppGpp-binding to the active site. A, overview of the allosteric pppGpp-binding site in BsRelQ (PDB entry 5DED) with relevant interacting side chains labeled (21). B, the same site in SaRelP with equivalent residues (as per the alignment) labeled. The positively charged Lys-21, Lys-25, and Arg-28 have been replaced by Ser-53, Ser-57, and Asp-60, respectively, thus changing the electrostatic behavior of the binding site. His-73 and His-74, which are unique to SaRelP, are shown in blue. C, activity of SaRelP (measured as production of ppGpp from GDP and ATP per enzyme/min) in the presence of increasing amounts of ppGpp. The calculated IC50 value is shown. D, same titration as in C but using pppGpp. Error bars represent S.D. of the turnover estimates determined by linear regression. Each experiment was performed at least three times. E, effect of model single-stranded mRNA (MF) on the enzymatic activity of EfRelQ (empty circles) and SaRelP (filled circles) measured as production of ppGpp from GDP and ATP per enzyme/min. Enzymatic assays were performed with 250 nm (62.5 nm tetramer) enzyme, 200 μm [3H]GDP, and 1 mm ATP. Error bars represent S.D. of the turnover estimates determined by linear regression; each experiment was performed at least three times. F, EMSA analysis of mRNA (MF) complex formation using increasing concentrations of EfRelQ and SaRelP. EMSAs were performed using 0.19 μm mRNA (MF) and increasing concentrations of SaRelP or EfRelQ as indicated on the figure.