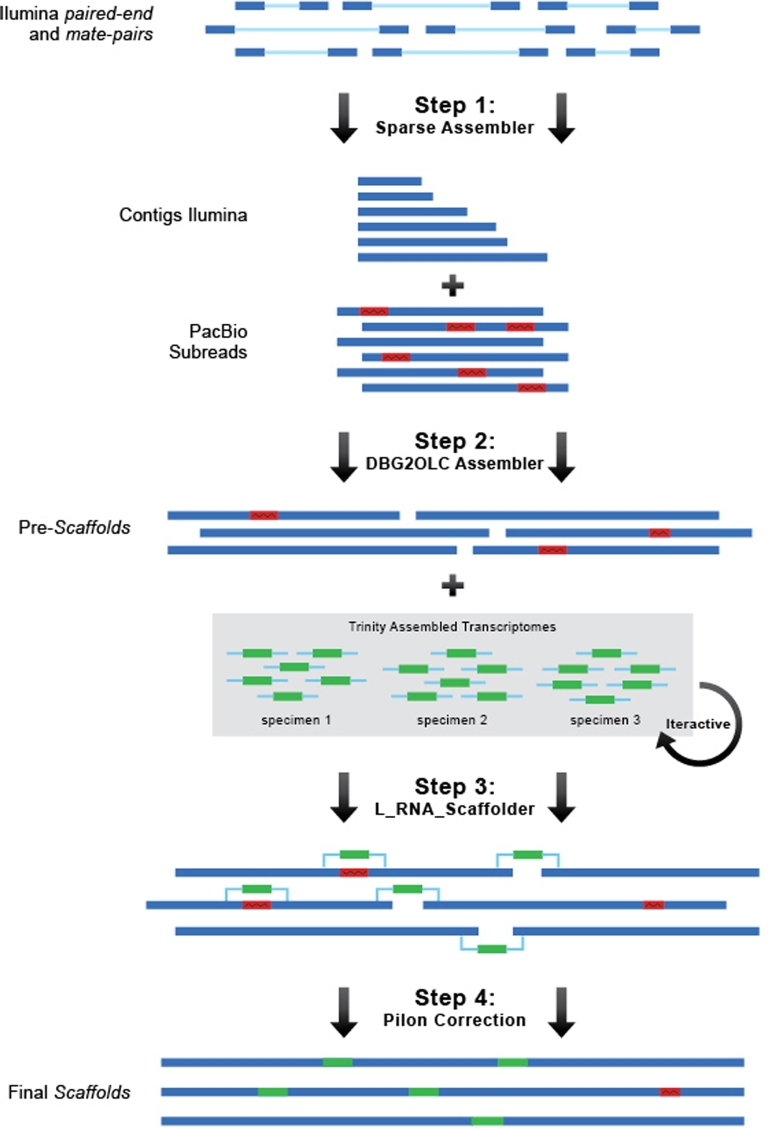
Figure 2:
Hierarchical assembly strategy employed for the golden mussel genome assembly. Trimmed Illumina reads were assembled to the level of contigs with the Sparse Assembler algorithm (Step 1). Then, Illumina contigs and PacBio reads were used to build scaffolds with the DBG2OLC assembler, which anchors Illumina contigs to erroneous PacBio subreads, correcting them and building longer scaffolds (Step 2), followed by transcriptome joining scaffolds using L_RNA_scaffolder (Step 3). Final scaffolds were corrected by re-aligning all Illumina DNA and RNA-seq reads back to them and calling consensus with Pilon software (Step 4). In bold is the bioinformatics software used in each step. Red blocks indicate PacBio errors, which are represented by insertions and/or deletions, found in approximately 12% of PacBio subreads.