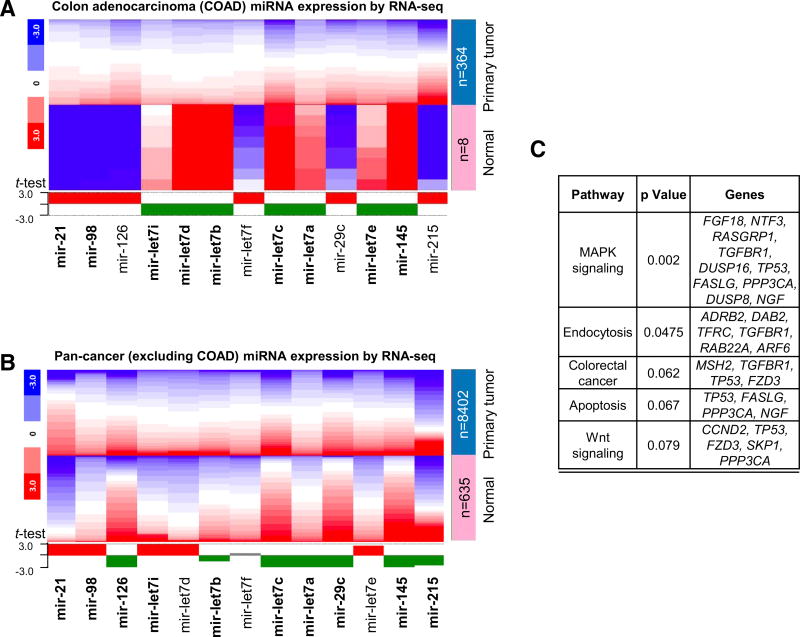
Fig. 4.
PhIP miRNA signature from rats recapitulated in human primary tumors. a TCGA data were mined for PhIP signature miRNA levels in human colon primary adenocarcinomas (COAD) compared with normal tissue. The heatmap is color-coded to indicate the relative gene expression level within each dataset. The expression profile of each miRNA that recapitulated the corresponding outcome in rat is shown in bold. b Corresponding data for pan-cancer, excluding the colon adenocarcinoma (COAD) data already analyzed in a. c Functional annotation of gene targets of PhIP signature miRNAs. Mir-21, mir-145, and let-7 family members were entered into TargetScan (http://www.targetscan.org), which identified 180 predicted target genes. These predicted targets were uploaded into DAVID, and the functional annotation results for the top five KEGG pathway terms were listed, as shown