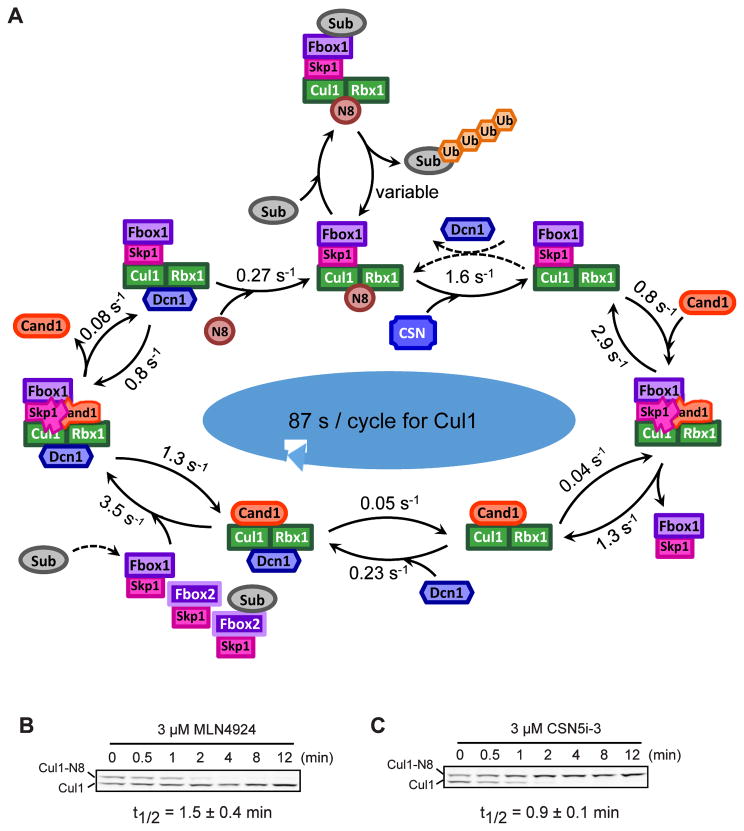
Figure 7. Rapid cycling of Cul1 in human cells.
(A) Cycling of Cul1 summarized from biophysical, cellular and computational studies. Association rates are computed based on kon and steady state cellular concentrations of unbound proteins, and the cycle time for Cul1 is computed using effective rates for the reversible binding steps (see also Fig S5D). The reversal of the de-neddylation reaction by Dcn1 (dashed lines) is discouraged in WT cells due to preferential association of Dcn1 with Cand1•Cul1, but is expected to occur more frequently in DKO cells. The substrate of the SCF complex can bind the FBP either in its free or assembled state. Substrate binding stabilizes the SCF complex by preventing CSN from binding. The 55 s cycle time for Cul1 represents the average time it takes a Cul1 molecule to be deneddylated and exchanged into a different SCF if it is not bound by substrate.
(B) Deneddylation of Cul1 is fast in human cells. HEK293 cells were treated with 3 μM MLN4924 to inhibit the Nedd8 E1 and were maintained at 37°C fo r the indicated time before being directly lysed on culture plates. Average t1/2 for deneddylation is shown. Error bars, ± SD, n = 3.
(C) Neddylation of Cul1 is fast in human cells. Assay condition was similar to (B) but 3 μM CSN5i-3 was used to inhibit CSN (Schlierf et al., 2016). Average t1/2 for neddylation is shown. Error bars, ± SD, n = 3.