Figure 5.
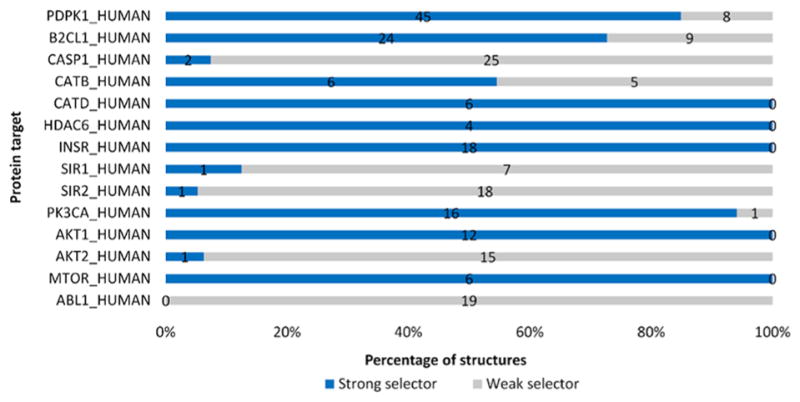
Percentage of the strong selector structures among 14 targets in the protein test-set. Crystal structures which have a good performance in distinguishing active ligands from inactive ligands are labeled as strong selectors. The number on the bar represents the quantity of strong or weak selector structures for each target. In total, 249 PDB structures in the structure test-set are examined while 142 structures are strong and 107 structures are weak. Among all the 14 targets in this test, 13 targets have at least one strong selector structure according to ProSelection. Abbreviations: PDPK1, 3-phosphoinositide-dependent protein kinase 1; B2CL1, Bcl-2-like protein 1; CASP1, Caspase-1; CATB, Cathepsin B; CATD, Cathepsin D; HDAC6, Histone deacetylase 6; INSR, Insulin receptor; SIR1, NAD-dependent protein deacetylase sirtuin-1; SIR2, NAD-dependent protein deacetylase sirtuin-2; PK3CA, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform; AKT1, RAC-alpha serine/threonine-protein kinase; AKT2, RAC-beta serine/threonine-protein kinase; MTOR, Serine/threonine-protein kinase mTOR; ABL1, Tyrosine-protein kinase ABL1.
