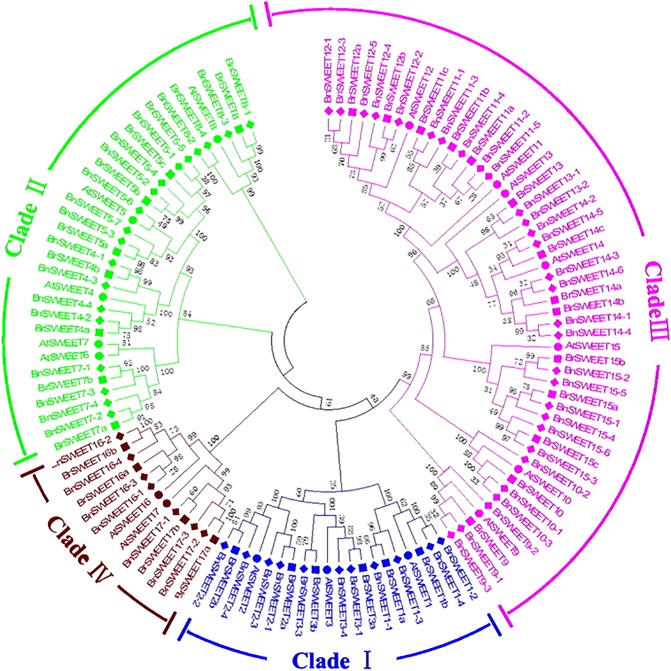
FIGURE 2.
Phylogenetic tree of SWEETs from B. rapa, B. napus, and A. thaliana. The phylogenetic tree was built using the neighbor-joining (NJ) method. The unrooted tree was generated using ClustalW in MEGA5 via SWEETs amino acid sequences from A. thaliana (AtSWEET), B. rapa (BrSWEET), and B. napus (BnSWEET). The roman numerals (I – IV) labeled with various colors indicate different clades. The numbers at the nodes represent bootstrap percentage values based on 1000 replications. Genes from each species are marked with different bullet point colors.