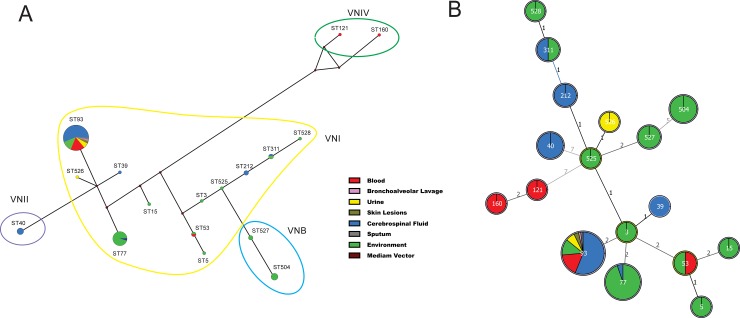
Fig 4.
Median-joining haplotype network (A) of 102 clinical and environmental C. neoformans isolates based on concatenated nucleotide sequences of the seven MLST loci. The major molecular types found in this study (VNI, VNII, VNIV, and VNB) are indicated by circles of different colours. Each circle represents a unique Sequence Type (ST), and the circumference is proportional to sequence type frequency (ST93: 77 isolates; ST77: 19; ST504: 5; ST40: 4; ST53: 2; ST527: 2; ST311: 2; ST212: 2; ST528: 1; ST526: 1; ST525: 1; ST160: 1; ST121: 1; ST39: 1; ST15: 1; ST5: 5; ST3: 1). Brown dots (median vectors) are hypothetical missing intermediates. Minimum spanning trees (B) using the goeBURST algorithm among C. neoformans isolates determined by median-joining network analysis. The size of the circle corresponds to the number of isolates within that haplotype, and the numbers between haplotypes represent the genetic distance of each haplotype, excluding the gaps. The figure shows the distribution of sequence types according to the clinical site and environmental origin.