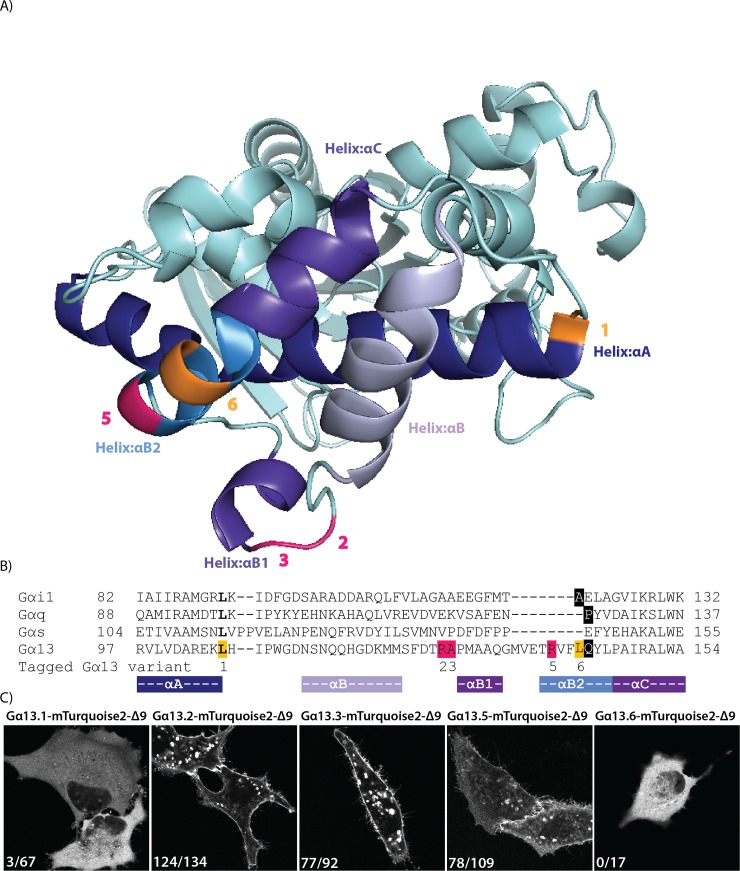
Fig 1. Insertion of a fluorescent protein at different positions in Gα13.
(A) The protein structure of human Gα13 (PDB ID: 1ZCB). The highlighted residues indicate the amino acid preceding the inserted fluorescent protein. Successful sites for inserting mTurquoise2-Δ9 into Gα13 in pink and unsuccessful sites in orange. (B) A partial protein sequence alignment (full alignment see S1 Fig) of different Gα classes. The highlighted residues indicate the amino acid preceding the inserted fluorescent protein (or luciferase). In bold, the sites that were previously used to insert Rluc [26]. Insertion of mTurquoise2-Δ9 in Gα13 after residue Q144 (black) was based on homology with previous insertions in Gαq and Gαi (black). Successful sites for inserting mTurquoise2-Δ9 (R128, A129 and R140) in pink and unsuccessful sites (L106 and L143) in orange. The numbers indicated below the alignment correspond with the Gα13 variant numbers, used throughout the manuscript. The colors under the alignment match with the colors of the αHelices shown in (A). (C) Confocal images of the tagged Gα13 variants transiently expressed in HeLa cells. The numbers in the left bottom corner of each picture indicate the number of cells that showed plasma membrane localization out of the total number of cells analyzed. The tagged Gα13 variants also localize to structures inside the cell, which are presumably endomembranes,. The width of the images is 76μm.