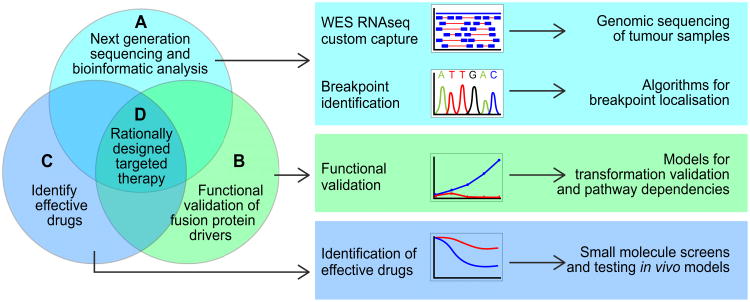
Figure 1. Methods used to identify, validate and target oncogenic fusion proteins.
(A) A variety of NGS methods generate data from primary patient samples and many different computational methods have been developed to identify fusion proteins (see also: Section – New opportunities for detecting fusion proteins and Table 2). These algorithms vary depending upon the type of genomic sequencing employed and whether the fusion is known or novel. Sanger or deep re-sequencing based confirmation of the fusion breakpoint is important for ruling out false-positive results. (B) Functional validation experiments must be carried out to determine how fusion proteins promote tumorigenesis. Rationally chosen cell line or animal model systems, created to test whether the fusion protein possesses transformative ability, can also be used to identify pathway dependencies. (C) These model systems can also be used to identify small-molecule inhibitors and clinically actionable drugs capable of blocking fusion protein function. (D) Together, these data support the clinical development of rationally designed targetted therapies for fusion protein-driven cancers.