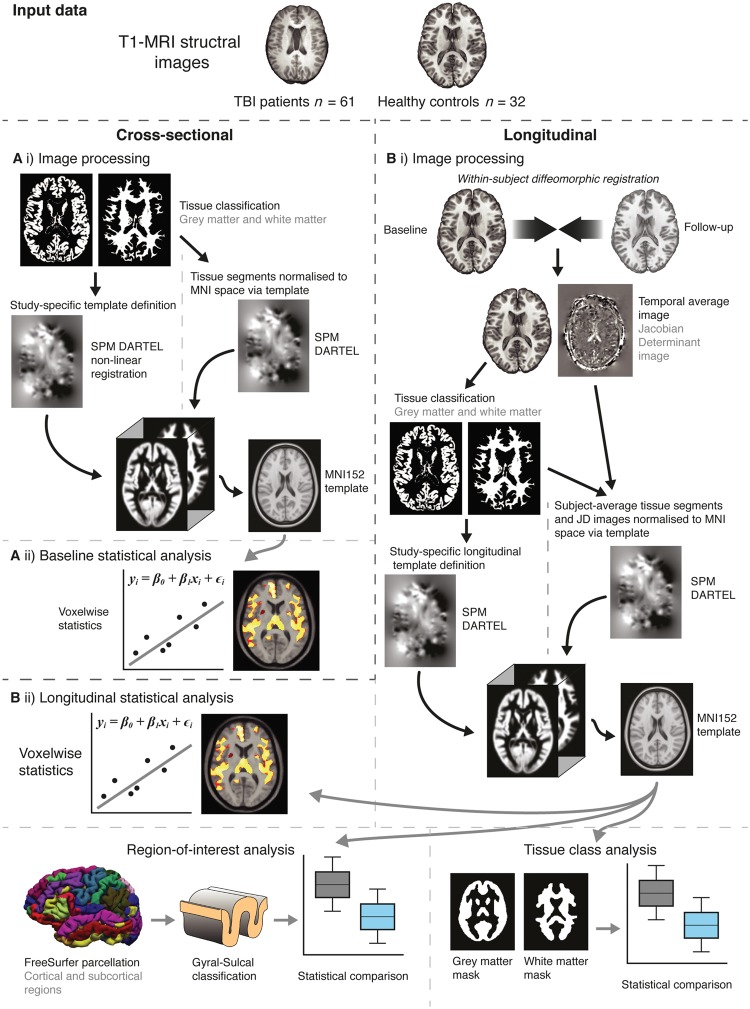
Figure 1.
Overview of study methods. [A(i)] Initial processing used SPM to segment T1 images into grey and white matter probability maps. A study-specific template was defined based on 40 randomly selected participants (20 TBI patients, 20 controls), using DARTEL registration for non-linear spatial normalization. The template was then affine registered to MNI152 space. All images were then normalized (smoothed by 8 mm and modulated) to MNI152 space via the study template. [A(ii)] Statistical analysis was carried out voxelwise across the normalized grey and white matter images, using Randomise (FSL) with 5000 permutations using the threshold-free cluster enhancement, adjusting for age and intracranial volume. [B(i)] Image processing entailed an initial symmetric within-subject registration for each subject’s baseline and follow-up images. This generated a within-subject ‘temporal average’ and Jacobian determinant image, representing the voxelwise spatial expansion and contraction necessary to match baseline and follow-up images. Average images were then segmented into grey and white matter. A random selection of 20 TBI patients and 20 controls was used to define a study-specific longitudinal template with DARTEL, which was then affine registered MNI152 space. Individual average images and Jacobian determinant images were then normalized (smoothed by 8 mm and modulated) to MNI152 space via the longitudinal template. [B(ii)] Longitudinal analysis included voxelwise group comparisons using Randomise, region of interest analysis based on FreeSurfer (Destriuex) atlas regions and tissue class (i.e. grey and white matter) analysis.