(
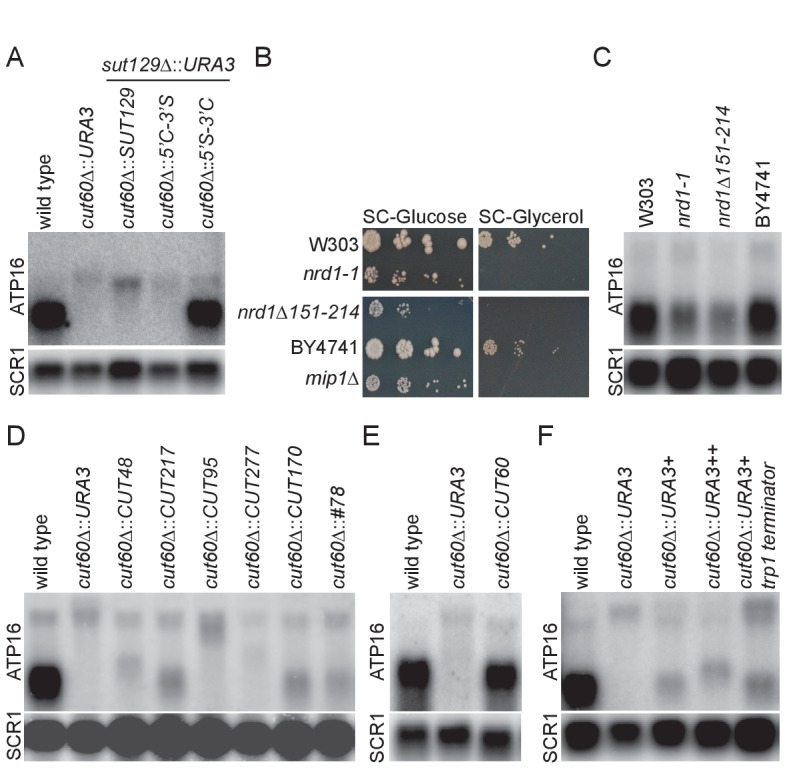
A) Schematic representation of the locus between
MED2 and
ATP16 when the 5’ sequence of
SUT129 is fused to the 3’ sequence of
CUT60, and this fusion sequence is inserted into the location of
CUT60. The DNA sequences of the individual parts are shown in the lower panel. (
B) Schematic representation of the locus between
MED2 and
ATP16 when the 5’ sequence of
CUT60 is fused to the 3’ sequence of
SUT129, and this fusion sequence is inserted into the location of
CUT60. The DNA sequences of the individual parts of this locus are shown in the lower panel. (
C) Quantification of
ATP16 expression in northern blot shown in
Figure 3A normalized to wild type. Data of expression in wild type,
cut60Δ
::URA3,
cut60Δ
::SUT129 + sut129Δ
::URA3,
cut60Δ
::5’C-3’S + sut129Δ
::URA3 and
cut60Δ
::5’S-3’C + sut129Δ
::URA3 strains. Error bars are s.e.m. of 4 biological replicates (
D) Northern blot analysis with a probe against the 5’-region of
SUT129 transcripts in wild type,
cut60Δ
::URA3,
cut60Δ
::SUT129 + sut129Δ
::URA3,
cut60Δ
::5’C-3’S + sut129Δ
::URA3 and
cut60Δ
::5’S-3’C + sut129Δ
::URA3 strains. For all strains the transcripts were analyzed in three different backgrounds: wild type,
rrp6Δ and
xrn1Δ indicated on top (n = 1). (
E) Serial dilution growth of W303 (wild type for
nrd1-1),
nrd1-1,
nrd1Δ151–214 and BY4741 (wild type for
nrd1Δ151–214) strains on SC-Ethanol plates (n = 2). (
F) Quantification of
ATP16 expression in northern blot shown in
Figure 3C.
ATP16 expression in W303 (wild type for
nrd1-1) and
nrd1-1 are normalized to
W303. ATP16 expression in BY4741 and
nrd1Δ151–214 are normalized to BY4741 (wild type for
nrd1Δ151–214). Error bars are s.e.m. of 3 biological replicates. (
G) Schematic representation of the three steps needed to generate precise site-specific genomic replacements of
CUT60. In step1 wild type yeast needs to be transformed with a PCR product containing the coding sequence of
URA3 with overhangs homologous to the sequences upstream and downstream of
CUT60. These transformants need to be selected on plates lacking uracil (SC-URA). In step two these transformants need to be transformed with a PCR product containing the sequence of the non-coding sequence for desired
CUT60 replacements (
CUT95 is shown as example) with overhangs homologous to the sequences upstream and downstream of
CUT60. In Step 3, transformants are selected on plates containing 5-FOA and additional uracil (SC + URA + 5 FOA) and confirmed by genotyping.