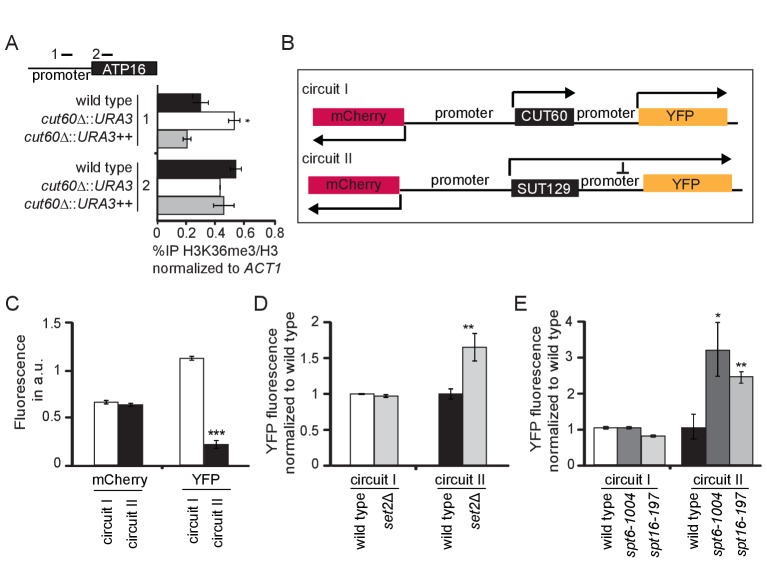
Figure 4. CUT60 prevents ATP16 repression by Transcriptional Interference.
(A) ChIP-qPCR experiments for histone H3K36 trimethylation (H3K36me3) relative to the histone H3 levels at the promoter and gene body of ATP16 in wild type (black), cut60Δ::URA3 (white) and cut60Δ::URA3++ (grey) strains. Error bars are s.e.m. of three biological replicates. * indicates p values of p<0.05 comparing %IP of H3K36/H3 in cut60Δ::URA3 to the %IP of H3K36me3/H3 in wild type. Position of ChIP-qPCR probes indicated in top panel. (B) Schematic representation of synthetic circuits. mCherry replaces MED2, YFP replaces ATP16. Circuit I represents wild type circuit containing CUT60, circuit II represents the cut60Δ::SUT129 mutant circuit. (C) Relative fluorescence of mCherry and YFP in circuit I (white) and circuit II (black). Error bars are s.e.m. of nine biological replicates, the mean was derived from multiple thousand individual measurements. *** indicates p values of p<0.0001. (D) Relative YFP fluorescence of circuit I in wild type (white) and set2Δ (light grey) background, and of circuit II in wild type (black) and set2Δ (light grey) background. YFP fluorescence levels are normalized to the corresponding wild-type values. Error bars are s.e.m. of six biological replicates. ** indicates p values p < 0.01 comparing YFP fluorescence in set2Δ to the respective wild type. (E) Relative YFP fluorescence in circuit I in wild type (white), spt6-1004 (dark grey) and spt16-197 (light grey) background, and in circuit II in wild type (black), spt6-1004 (dark grey) and spt16-197 (light grey) background. YFP fluorescence is normalized to the corresponding wild-type values. Error bars are s.e.m. of six biological replicates. * and ** indicate p values of p<0.05 and p<0.01 respectively comparing YFP fluorescence in spt6-1004 and spt16-194 to the respective wild type.
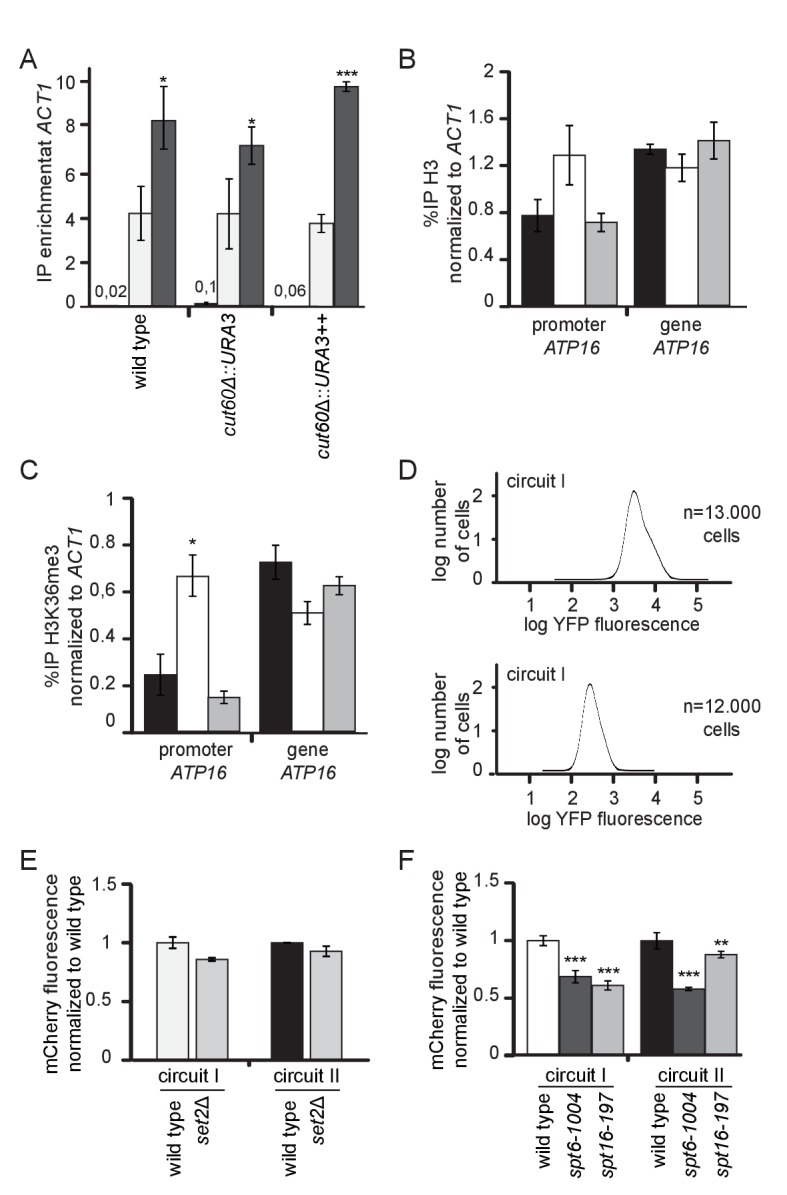
Figure 4—figure supplement 1. Set2p, Spt6p and Spt16p participate in Transcriptional Interference of ATP16.