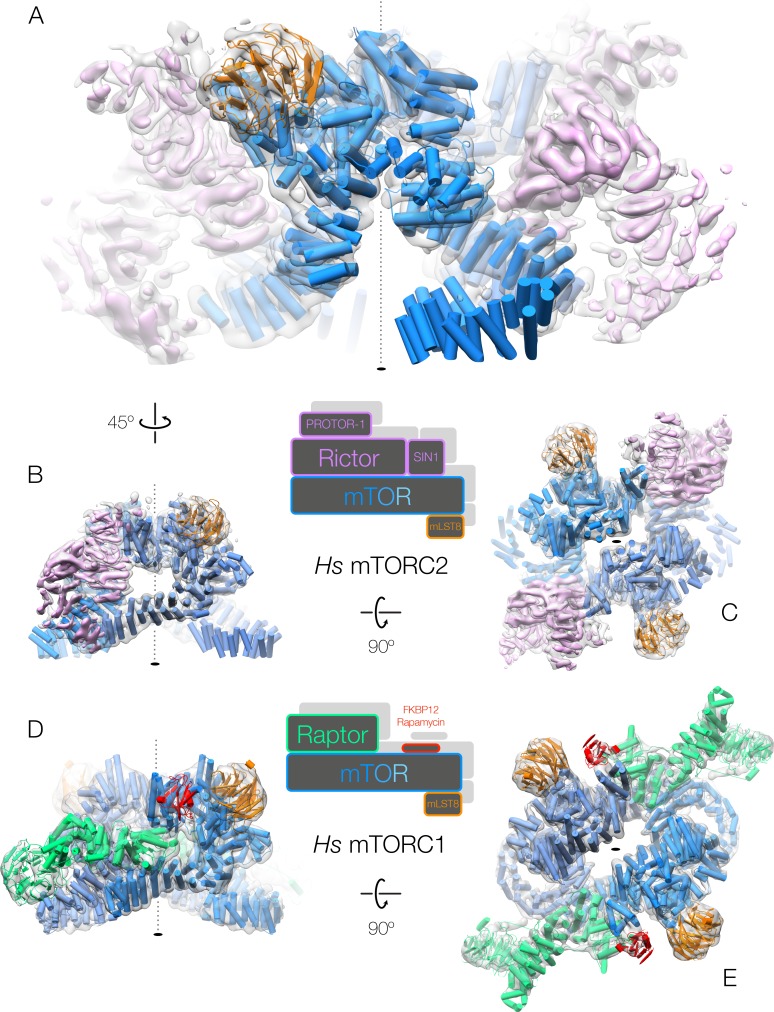
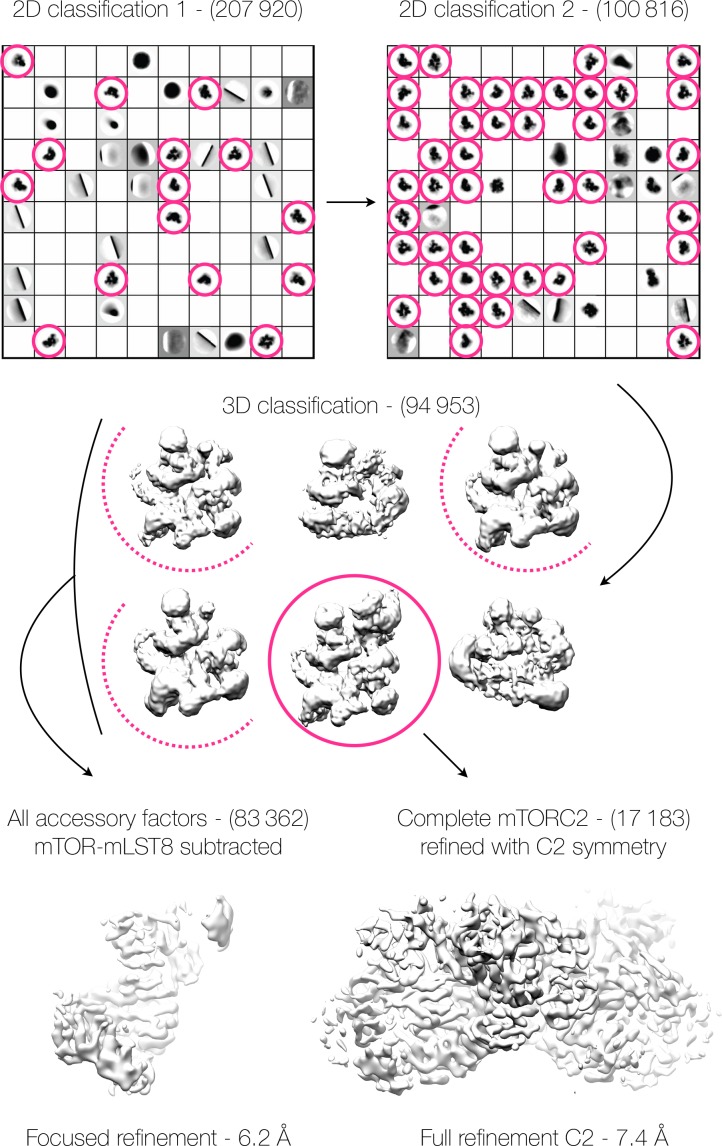
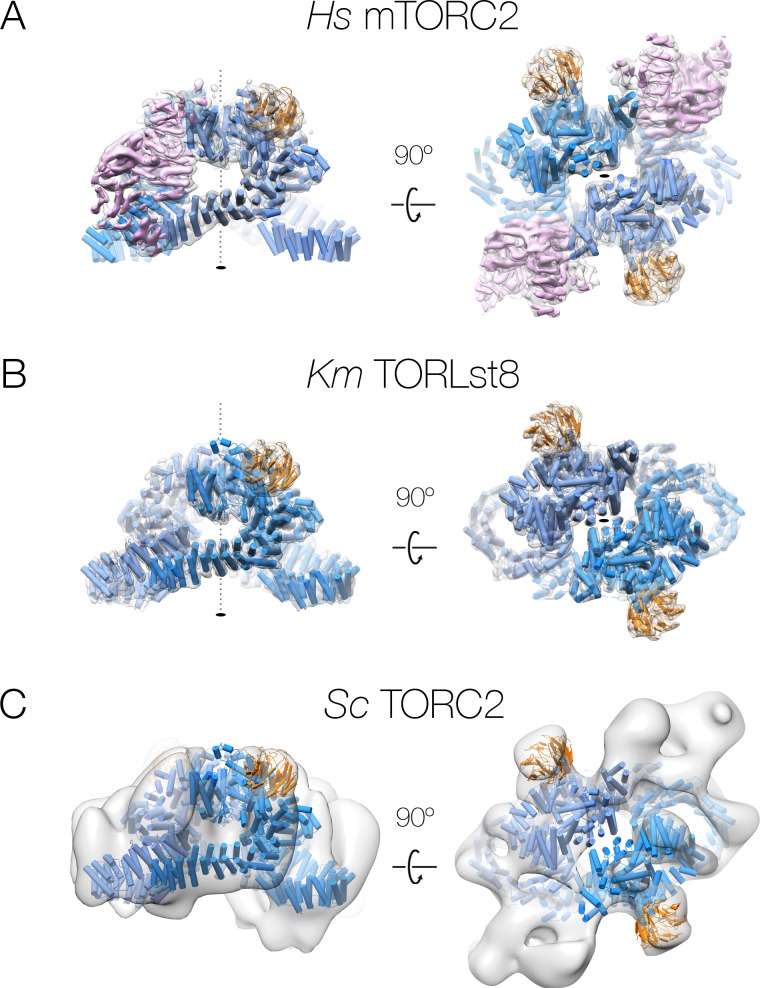
Figure 1. Both human mTOR complexes resolved at intermediate resolution.
(A–C) The architecture of human mTORC2. The structure is shown rotated as indicated by the arrows between the panels. The accessory factor density from focused refinement is shown within the dimeric, C2-symmetric mTORC2 density in pink. (D–E) The architecture of human mTORC1 (Aylett et al., 2016). The structure is shown rotated as indicated by the arrows between the panels. All complexes are shown with cryo-EM density as a grey transparent surface and the fitted structures in cartoon representation, coloured according to the primary structure schematics shown between the corresponding panels.