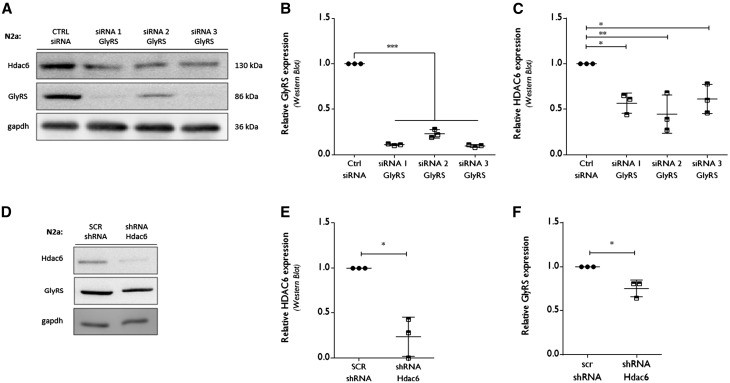
Figure 2.
RNA interference-mediated knock down of GlyRS affects HDAC6 expression. (A) N2a cells were transfected with siRNA molecules directed against Gars (GlyRS) mRNA or a control siRNA. At 72 h post transfection, cells were collected to check protein expression levels by western blot analysis. GAPDH was used as a loading control. (B) Using densitometry, the GlyRS protein expression level was determined, relative to GAPDH. All values were normalized to control siRNA samples within each experiment: control siRNA 1.00 ± 0.00, n = 3 versus siRNA 1 GlyRS 0.11 ± 0.01, n = 3 versus siRNA 2 GlyRS 0.23 ± 0.05, n = 3 versus siRNA 3 GlyRS 0.09 ± 0.02; one-way ANOVA, F(3,8) = 1.935, P < 0.0001. (C) The HDAC6 expression levels were analysed using densitometry, relative to the gapdh expression levels and normalized to control siRNA values: control siRNA 1.0 ± 0, n = 3 versus siRNA 1 GlyRS 0.57 ± 0.11, n = 3 and siRNA 2 GlyRS 0. 45 ± 0.21, n = 3 and siRNA 3 GlyRS 0.61 ± 0.16, n = 3; One-way ANOVA, F(3,8) = 8.327, P = 0.0076; Dunnett’s multiple comparisons test. (D) pCMV-dsRed plasmids containing either a scrambled shRNA or short hairpin RNA (shRNA) directed against Hdac6 mRNA were transfected in N2a cells. At 72 h post transfection, cells were collected to check protein expression levels by western blot analysis. (E) HDAC6 expression was checked by western blot analysis after 72 h shRNA-mediated knockdown. Densitometry was used to calculate HDAC6 expression levels, relative to GAPDH and all values were normalized to scrambled (SCR) shRNA samples: scrambled shRNA 1.00 ± 0.00, n = 3 versus shRNA Hdac6 0.24 ± 0.22, n = 3; one-sample t-test, t = 6.088, P = 0.0259. (F) The relative GlyRS expression levels were calculated using densitometry, relative to GAPDH levels and normalized to scrambled shRNA values: scrambled shRNA 1.0 ± 0.0, n = 3 versus shRNA Hdac6 0.75 ± 0.09, n = 3; one sample t-test, t = 4.353, P = 0.0489.