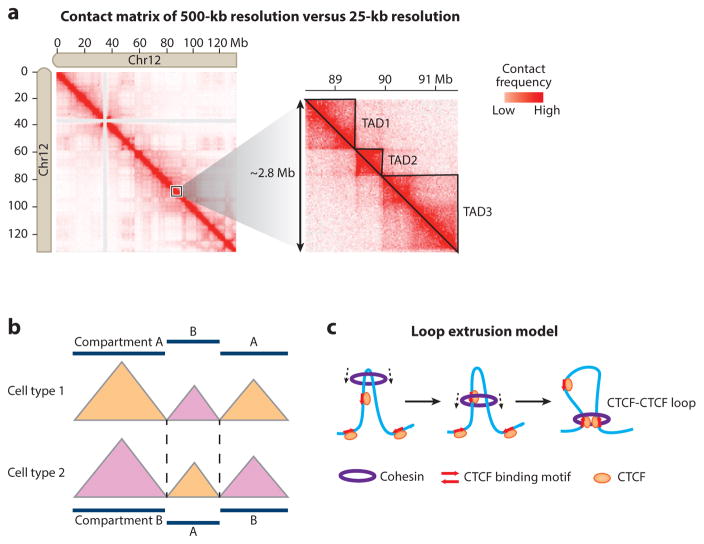
Figure 4.
Identification and characterization of topologically associating domains (TADs). (a) (Left) Cis-contact matrix for chromosome 12 from human embryonic stem cells at 500-kb resolution. (Right) A detailed contact matrix of an ~2.8-Mb region at 25-kb resolution reveals TAD organization as triangles. TAD1–3 are manually annotated and indicated by solid lines. The Hi-C data are taken from Dixon et al. (2012) and are visualized with Juicebox (Durand et al. 2016). (b) The position of TAD boundaries is largely conserved between different cell types, but TADs may transit between different compartments. (c) TAD formation by loop extrusion. The Cohesin complex forms a ring structure and travels along the DNA fiber, extruding a progressively larger loop until it is stalled by bound CTCF with convergent orientation (red arrows).