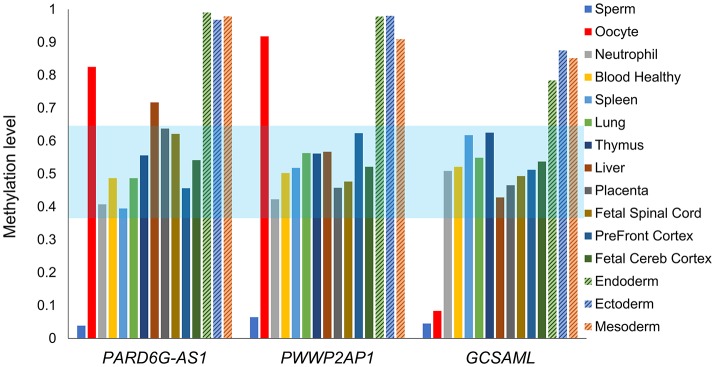
Figure 5.
Intermediate mean methylation level across the PARD6G-AS1, PWWP2AP1, and GCSAML candidate iDMRs. Comparative variation in the average methylation levels across all 5mCpG sites on the candidate iDMRs in the methylomes of healthy tissues depicted in Figures 2–4. In adult somatic tissues, the overall mean levels were 0.39 (PARD6G-AS1 iDMR), 0.42 (PWWP2AP1 iDMR) and 0.42 (GCSAML iDMR). A slightly skewed hypermethylation state (0.70) was observed in the liver for the PARD6G-AS1 iDMR. The three candidate iDMR regions are consistently hypermethylated in hESC-derived CD56+ ectoderm and mesoderm and hESC-derived CD184+ endoderm cell cultures. The light blue zone represents the expected intermediate methylation range (0.35–0.65). Each methylome is represented by a different color of bar.