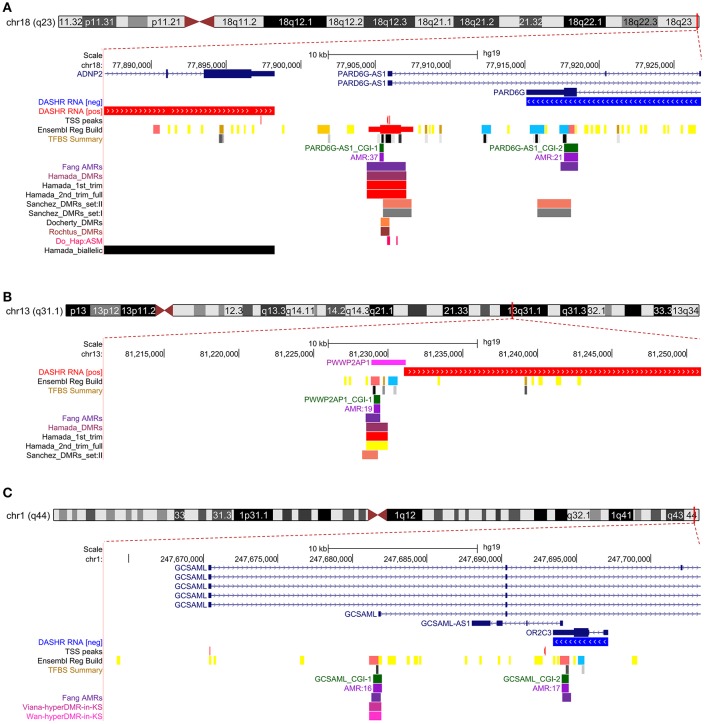
Figure 6.
Cross-reference overlaps of the PARD6G-AS1, PWWP2AP1, and GCSAML candidate iDMRs with differentially methylated regions reported in control-disease methylomes and healthy placentas. Customized graphics of the intersections between the candidate iDMRs at PARD6G-AS1 (A), PWWP2AP1 (B) and GCSAML (C) and the public records reporting parent-of-origin predictions, abnormal methylation profiles, and haplotype-dependent allele-specific 5mCpG sites. In each panel, the annotated features are (from top to bottom) the exon-intron organization of the principal and alternative isoforms, the evidence of large intergenic non-coding RNAs (lincRNAs) and small non-coding RNAs (sncRNAs) (DASHR RNA [pos] and [neg] tracks), the FANTOM5 transcriptional start sites (TSS peaks track) (Lizio et al., 2017), the Ensembl Regulatory Build predicted promoters (Ensembl Reg Build track) and transcriptional factor binding sites (TFBS summary track) (Cunningham et al., 2015), CpG islands and CGI-bearing AMRs (this study) and the comparative custom Fang_AMR track (Fang et al., 2012). (A) The PARD6G-AS1 CGI-1 overlaps the domain previously predicted to be differentially methylated in a parent-of-origin-dependent manner in blastocysts and placenta (Okae et al., 2014; Hamada et al., 2016; Sanchez-Delgado et al., 2016a) (Hamada_DMRs, Sanchez_DMRs_set:I, and Sanchez_DMRs_set:II tracks, respectively), for which the parental origin of the methylated alleles had not been established. The allelic methylation levels in the aforementioned regions are maternally skewed, varying from 0.69 in first- and second-trimester to 0.82 in full-term placentas (Hamada_1st_trim and Hamada_2nd_trim_full tracks). The PARD6G-AS1 CGI-1 also intersects with the domains reported to be hypomethylated in patients with Beckwith-Wiedemann syndrome and transient neonatal diabetes (Docherty et al., 2014) (Docherty_DMRs track) and in pseudohypoparathyroidism patients with GNAS cluster imprinting defects (Rochtus et al., 2016) (Rochtus_DMRs track), as well as with the haplotype-dependent allele-specific 5mCpG sites reported in placenta, liver, lung, brain and T cells (Do_Hap:ASM track) (Do et al., 2016). The genes that are known to be biallelically expressed in placenta (Hamada et al., 2016) are depicted in the Hamada_biallelic track. (B) The PWWP2AP1 CGI-1 overlaps the domain previously predicted to be differentially methylated in a parent-of-origin-dependent manner in blastocysts and placenta (Hamada et al., 2016; Sanchez-Delgado et al., 2016a) (Hamada_DMRs, and Sanchez_DMRs_set:II tracks, respectively), for which the parental origin of the methylated alleles had not been established. The methylation levels in the aforementioned regions varied from 0.38 in first- and second-trimester placentas to 0.24 in full-term placentas (Hamada_1st_trim and Hamada_2nd_trim_full tracks). (C) The GCSAML CGI-1 overlaps the domain reported to be skewed toward hypermethylation in Klinefelter syndrome (47,XXY karyotype) versus control males (Viana et al., 2014; Wan et al., 2015) (Viana-hyperDMR-in-KS and Wan-hyperDMR-in-KS), for which the parental origin of the allele methylation had not been established.