Figure 7.
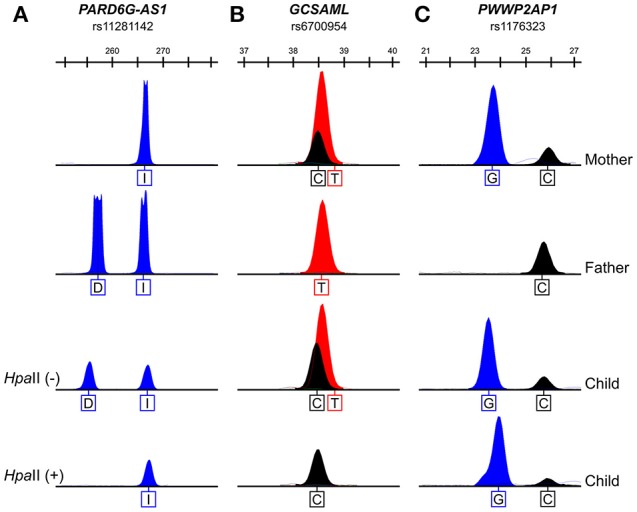
The 5mCpG sites at the PARD6G-AS1 and GCSAML iDMRs are on the maternally transmitted alleles. The maternally transmitted alleles and not the paternally transmitted alleles at the PARD6G-AS1 (A) and GCSAML (B) DMRs are consistently methylated, as supported by the resistance of the maternally inherited alleles to digestion with the methylation-sensitive HpaII restriction enzyme and the lack of amplification of the paternally transmitted alleles from the digested genomic DNA samples for the informative variant SNPs (the PARD6G-AS1 upstream gene indel variant rs11281142 (I = insertion of CTGTGGTGC; D = deletion of CTGTGGTGC) and the GCSAML intron variant/upstream gene variant rs6700954, respectively) in three representative nuclear families that were informative for at least one SNP. Therefore, the 5mCpG marks in the PARD6G-AS1 and GCSAML DMRs are maternal imprints. On the other hand, both parental alleles of the PWWP2AP1 rs1176323 variant are resistant to digestion (C). Thus, the 5mCpG marks in the PWWP2AP1 CGI-1 occur in a parent-of-origin-independent manner. Given the intermediate rate of methylation in PWWP2AP1 CGI-1 (see Figure 5), the occurrence of 5mCpG sites in both parental alleles indicates that the PWWP2AP1 CGI-1 alleles are methylated in a stochastic fashion.
