Fig. 5.
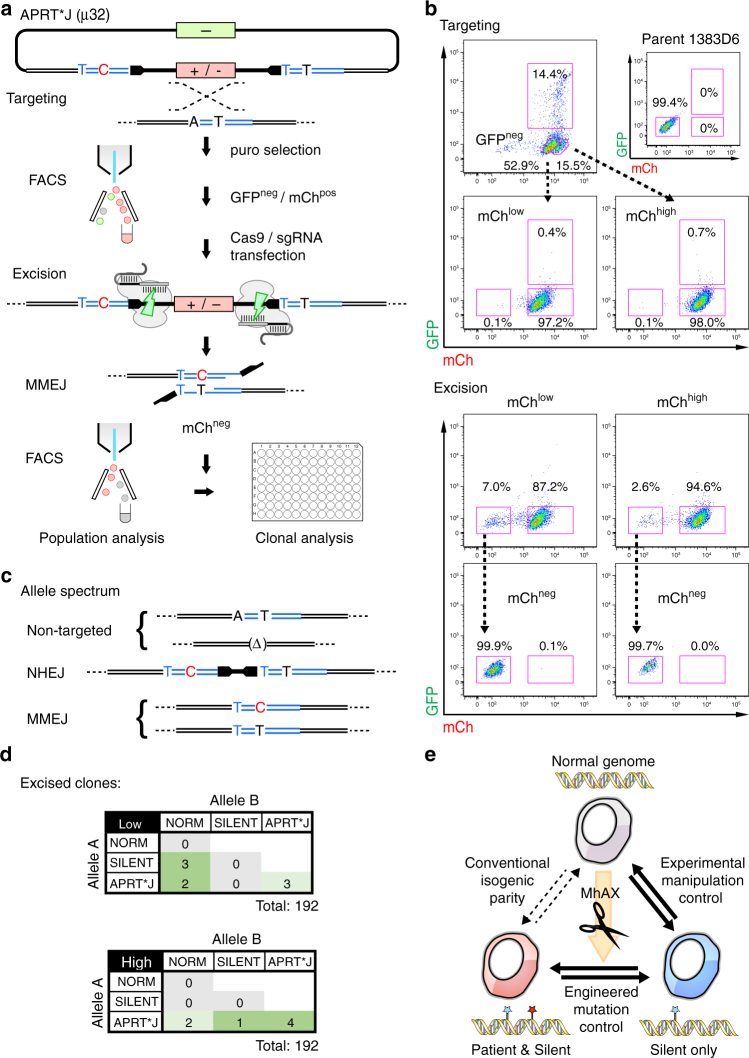
Expedited biallelic gene editing by FACS sorting. a Schematic of the FACS sorting protocol. GFPneg / mChpos cells (targeted) are isolated in bulk and subjected to nuclease transfection, followed by sorting populations for mChneg (excised). Resolved alleles were screened in the population, or in single-cell derived iPSC clones. The donor vector, allele and additional features are as described in Fig. 4a,b. b Representative FACS plot for the targeting and excision steps. GFPpos cells were excluded, and mChpos cells were divided into high and low fractions to bias monoallelically and biallelically targeted cells, respectively. c Predicted APRT allele spectrum within the sorted mChneg populations. d APRT diploid genotypes of clones. Only clones with Normal or MMEJ-resolved alleles are shown. Alleles marked as ‘APRT*J’ were also edited with the Silent mutation. e By employing imperfect μH, MhAX derives edited cells and their concordant isogenic controls simultaneously, reducing technical variation